Negative design to prevent evolutionary failure
Contributed by:



TYLER CAMP, DENNIS MISHLER, JEFFREY BARRICK, BARRICK LAB, Center for Systems and Synthetic Biology, Department of Molecular Biosciences, The University of Texas at Austin
Editor's note: When engineers build a bridge, they must design it to withstand heavy traffic, high winds, and earthquakes so that it will not fail when stressed. Genetic engineers face a very different kind of design challenge: unwanted evolution.The Barrick labat UT Austin discussed with Benchling tricks for overcoming this design challenge.
The Barrick labhas supported iGEM teams at UT Austin since 2012. They find the creativity of iGEM students inspiring. At UT Austin, the team has worked on projects—such as engineering caffeine-addicted E. coli—that have led to scientific publications. They hope that their work on evolutionary stability will help iGEM teams be successful in their most ambitious endeavors. They welcome feedback on these procedures and encourage teams to document the genetic stability of BioBricks on the iGEM Registry website.
At Benchling, we are dedicated to helping create and communicate cutting-edge research. If you want a similar site for your research, just contact us.
What is unwanted evolution?
Whenever we place a DNA sequence inside of a cell, there is a chance that it will mutate in a way that destroys or damages the information in our carefully crafted DNA program. If even a few cells in a population malfunction, these mutants can take over and become numerically dominant in the population in short order, especially if they grow more quickly as a result of circumventing our instructions.
How do we quantify stability against unwanted evolution?
We use the term "evolutionary stability" to refer the ability of a DNA sequence to continue to function over the course of many cell divisions. A common way to measure the evolutionary stability of a genetic device is as a "half-life" of its functional decay. For example, in a genetic circuit with fluorescent output, we would record the time (often measured in generations of cellular replication) it takes until we observe only 50% of the original fluorescent signal per cell. Loss of fluorescence in this case is due to mutations in the sequence required for producing the fluorescent protein.
One way to measure the evolutionary stability of a genetic device in a microbe is to pick an entire single colony that is derived from a fully fluorescent cell, propagate all of the cells in liquid media overnight, and follow with continued subculturing (diluting the culture into fresh media each day). The number of cells in a culture and the dilution factor up to that point can be used to estimate the number of generations that have elapsed from the initial single cell on the agar plate to the cells in the current culture. The level of function (fluorescence) remaining in a cell population can be monitored by plating a dilution of the culture to visualize many colonies on agar, by using a fluorometer, or by flow cytometry. By conducting this entire procedure with evolutionarily independent replicates, propagating and monitoring 6 to 12 cell populations derived from different initial colonies, one can estimate the half-life with which the function of this genetic device is lost due to mutations.
Here is a sample protocol we use in the lab:
What is a typical evolutionary half-life for a DNA device in an E. colicell?
University of Texas at Austin undergraduates involved in our Freshman Research Initiative stream studying synthetic biology and our 2015 iGEM team tested the half-lives of several plasmids expressing fluorescent proteins using various constitutive promoters and ribosome binding sites with different strengths.
These simple devices were constructed from BioBricks available from the iGEM Registry of Standard Biological Parts. For some—like the ones shown in Figure 1 —significant fluorescence was lost in many cells in a population by the time an initial 5 ml culture from the picked colony reached saturation (~35 generations), and function was entirely lost after the first day of subculturing (~45 generations) ( Figure 2 ). For others, full fluorescence was maintained over more than seven days of subculturing.
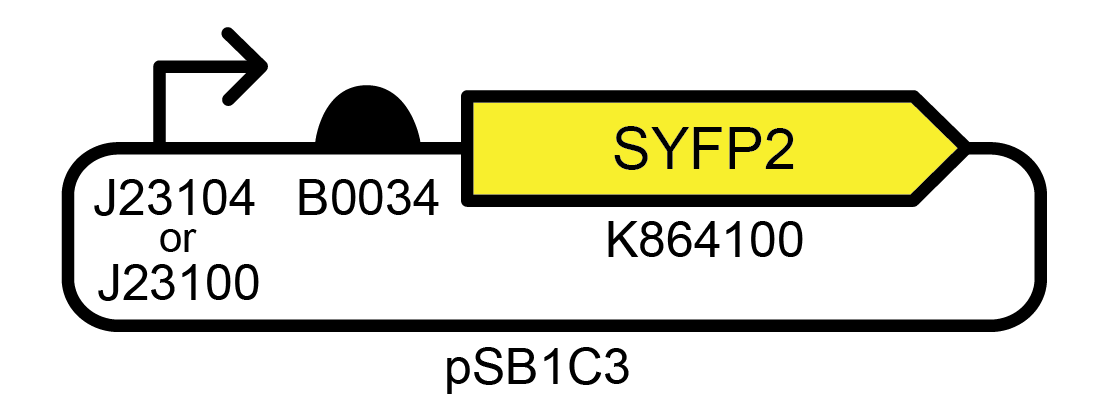
Figure 1. Example of an unstable genetic device. This high copy number E. coli plasmid strongly expresses a yellow fluorescent protein. Accession numbers for promoter, ribosome binding site, and fluorescent protein BioBricks are from the iGEM Registry of Standard Biological Parts. Profiling the evolutionary stability of two variants of this device with different promoters (J23104 or J23100) yielded very similar results.
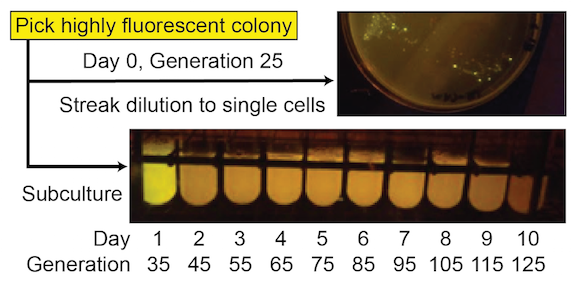
Figure 2. SYFP2 function from this device has a very short evolutionary half-life. Cells with mutated plasmids, which no longer express this fluorescent protein, reach a high frequency even during the initial growth of a colony on a plate (mixture of bright and nonfluorescent cells in the upper image). Fluorescence is entirely lost in all cells in the population after one additional day of growth after subculturing (loss of fluorescence in day 2 test tube culture in the lower image).
What happened to the "broken" variants of each plasmid?
If we were in the business of building bridges, at this point we would analyze our failures and then go back to the drawing board to figure out how to make more robust designs. So, these students sequenced "broken" variants of each plasmid isolated at the end of these evolution experiments. Most plasmids had a dominant type of mutation that reproducibly led to their failure, many times and in many independent cultures.
For the devices shown in Figure 1 , fluorescence was lost in a majority of cases due to a selfish mobile element from the E. coli genome (a Tn10 transposon) inserting a copy of itself near the beginning of the fluorescent protein open-reading frame in the plasmid ( Figure 3 ). This mutation disrupts expression of the fluorescent protein, apparently ameliorating the cost associated with this function. Base substitutions or a small deletion were observed in the promoter, start codon, and reading frame in other populations. These may similarly reduce or eliminate costly fluorescent protein expression.
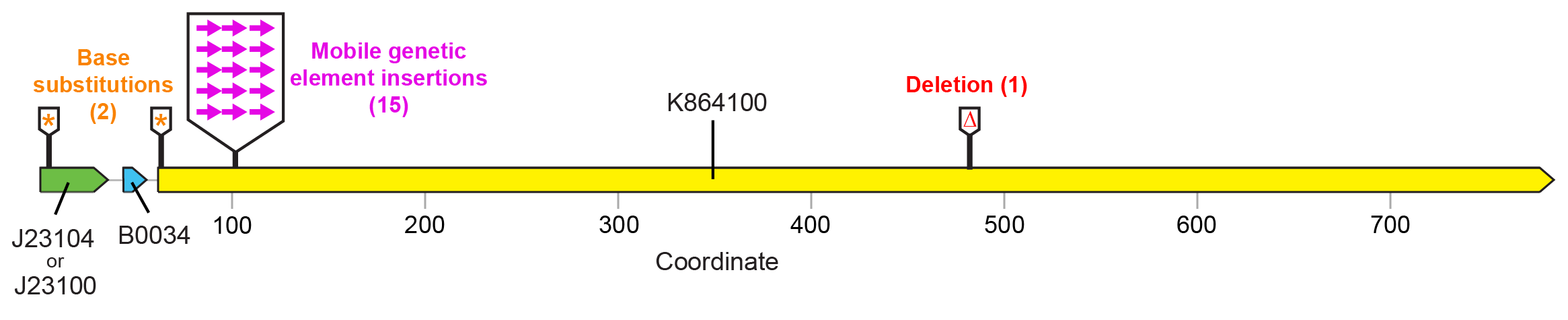
Figure 3. Failure modes found by sequencing "broken" SYFP2 plasmids. Nearly all (15/18) mutations that resulted in loss of fluorescence were due to insertions of a mobile genetic element at a specific target site located near the beginning of the SYFP2 reading frame. Editing the device sequence to remove this site would be expected to increase the stability of fluorescent protein expression because it would completely eliminate this very common type of failure mutation. The number of symbols at each location indicates how many of those mutants were recovered from separately evolved E. colipopulations.
What if we now redesigned the sequence of our device to edit out the "fragile" sequences that led to major "failure modes"? Would we be able to extend the useful lifetime of these devices? A pioneering study led by Dr. Sean Sleight in the research group of Prof. Herbert Sauro at the University of Washington showed that it is possible to achieve greater evolutionary stability using this strategy [1].
In the case of our example device, the instability caused by site-specific insertion of a mobile genetic element was very specific to the E. coli TOP10 host strain that was used in these experiments—this mobile genetic element is not even present in other strains. So, it might be avoided either by deploying the device in a different E. coli strain or by editing the sequence of the insertion site so that this element could no longer insert itself there.
How do you design robust DNA sequences?
Our lab has created a web tool, the Evolutionary Failure Mode (EFM) Calculator [2], to predict evolutionary weaknesses in a DNA sequence that are expected to operate in a wide variety of host organisms. By adding this in silico tool to a design-build-test cycle, one can rule out DNA sequences that are likely to be unstable over evolutionary time at the design phase.
Currently, the EFM Calculator predicts two types of well-characterized mutational hotspots that can be recognized in the primary sequence of a device: RMDs (repeat-mediated deletions) and SSRs (simple sequence repeats). For the first type (RMDs), repeated sequences, such as two copies of the exact same gene or even two copies of a short regulatory sequence, are prone to DNA recombination events that will delete any important sequences between the repeats. For the second type (SSRs), DNA polymerases often slip on tandem repeats of one or a few nucleotides (e.g., AAAAAAA or GACGACGACGAC), leading to deletions or insertions of bases (e.g., +A or ΔGAC) that may frameshift a protein or otherwise inactivate a device.
An example of EFM Calculator output for a popular BioBrick part from the iGEM registry is shown in Figure 4 . The EFM Calculator predicts a relative mutation rate for each example of a potentially unstable sequence based on its properties (e.g., size of repeat, distance between repeats). Then, these values are combined into an overall relative instability prediction (RIP) score for the entire DNA sequence.
When no hotspots are present, the RIP score is one. Greater RIP scores mean that the mutation rate could potentially be reduced by that factor if all of the predicted high-risk hotspots for mutations were edited out of its sequence and only the basal rate of base pair substitution (BPS) mutations remained. Thus, the EFM Calculator operates as a negative design tool for avoiding evolutionarily problematic sequences, much like a primer analysis program would predict hairpins, dimers, or nonspecific annealing that might interfere with PCR amplification.
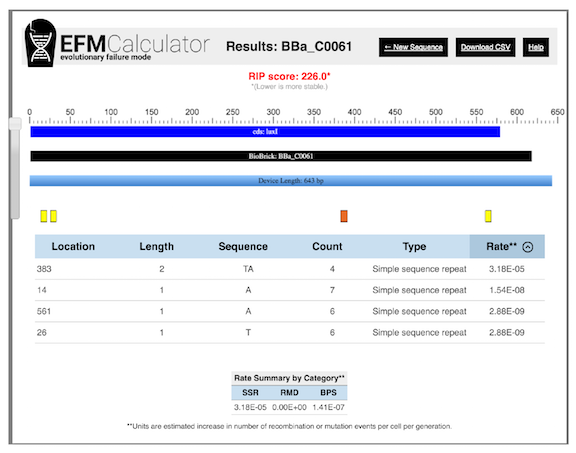
Figure 4. EFM Calculator results for a popular DNA part encoding a The relative instability prediction (RIP) score is an estimate of device instability, with lower scores representing more stable devices. In this case, it predicts that the mutation rate in the device could be reduced by a factor of 226 by editing out three simple sequence repeats that are likely to be hotspots for small insertion and deletion mutations. The longest repeat, which has the sequence TATATATA and is located at position 383, is predicted to contribute the most, by far, to the mutation rate in this device.
Conclusion
Improving evolutionary stability by minimizing mutation rates—and integrating this approach with strategies for reducing the fitness cost or metabolic load of genetic devices on host cells-can have many practical benefits. For example, if we metabolically engineer a strain to produce a drug or biofuel, it pays to not have cheater cells lurking in a fermenter that have mutated such that they no longer participate in creating the desired product. Evolution is also inherently random, so minimizing its influence on a system makes biology more amenable to predictive modeling.
As genetic engineers build ever-larger DNA programs with functions that are more complex and costly to host cells, it will become increasingly difficult to prevent these ambitious designs from collapsing under the pressure of evolution. By carefully analyzing these stresses, we're confident that future engineers will be able to stably "bridge" this gap.
For a full explanation of the EFM calculator, please see our full manuscript in ACS Synthetic Biology.
Don't forget to check out our online EFM calculator tool.
References
Sleight, S.C., Bartley, B.A., Lieviant, J.A., Sauro, H.M. (2010) Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng.4:12. PMID:21040586
Jack, B.R., Leonard, S.P., Mishler, D.M., Renda, B.A., Leon, D., Suárez, G.A., Barrick, J.E. (2015) Predicting the genetic stability of engineered DNA sequences with the EFM Calculator. ACS Synthetic Biol. 4:939-943. PMID:26096262
Acknowledgements
We thank the students involved in the University of Texas at Austin Freshman Research Initiative (FRI) Stream "Hijacking Microbial Factories for Synthetic Biology" and the 2015 University of Texas at Austin iGEM team. Research in the Barrick Lab on evolutionary stability is funded by the DARPA BRICS program (HR0011-15-C0095), an NSF CAREER grant (CBET-1554179), and the NSF BEACON Center for the Study of Evolution in Action (DBI-0939454).