Benchling Product Bulletin: Summer 2021
As we continue to extend the Benchling R&D cloud, we’re focused on meeting the needs of teams throughout the entire R&D lifecycle. In our most recent product update, we released new functionality for managing processes across R&D. This will be especially important to development teams, such as bioprocess and bioanalytical development, who require more process-driven workflows.
Create Structured Templates for R&D Processes in Benchling Notebook
Benchling now offers new fill-in template capabilities to create and lock down key processes that guide how teams complete tasks. Balancing flexibility with control, these templates provide teams the ability to configure Notebook to more regimented processes, such as repeatable experiments, analytical tests, QC protocols, and more.
How this feature helps you
Easy-to-follow fill-in templates help guide teams through their work, simplifying a wide range of daily tasks. This structure also reduces the potential for inadvertent data modifications. Team leads will have greater confidence that groups are entering data and following processes with compliance. This reduces the overall time needed to enter and review data, accelerating process execution end-to-end.
Key functionality
With these templates, you can:
Design, document, and lock down templates in Notebook
Control how teams fill in template information by leveraging three structured actions: entering data in a table, checking a checkbox, and uploading a file attachment
Restrict users from editing or deleting important entry information, such as protocol directions or data
Use cases
Groups across R&D can leverage these templates for a multitude of use cases, such as:
Defining and standardizing repeatable experiments in early discovery
Establishing structured assays and results capture in analytical testing
Locking down rigid, compliant protocols for sample testing in quality control
The latest Benchling release also delivers a number of helpful features across all our cloud applications. Learn more about these features by clicking each of the links below:
Notebook
Benchling Notebook’s easy-to-use interface lets you log experiments and protocols, track information, and record sample results, all while driving unified data capture across Benchling. In addition to new template capabilities detailed above, we’ve also added support for duplicating templates, along with enhanced table functionality and formulas.
Clone templates in Benchling Notebook
You can now duplicate entire Notebook templates — in addition to individual entries — and store these files in any template collection folder. When combined with more structured templates, template cloning accelerates the creation of multiple locked templates, which can be used across a variety of processes such as SOPs, protocols, and repeatable experiments.
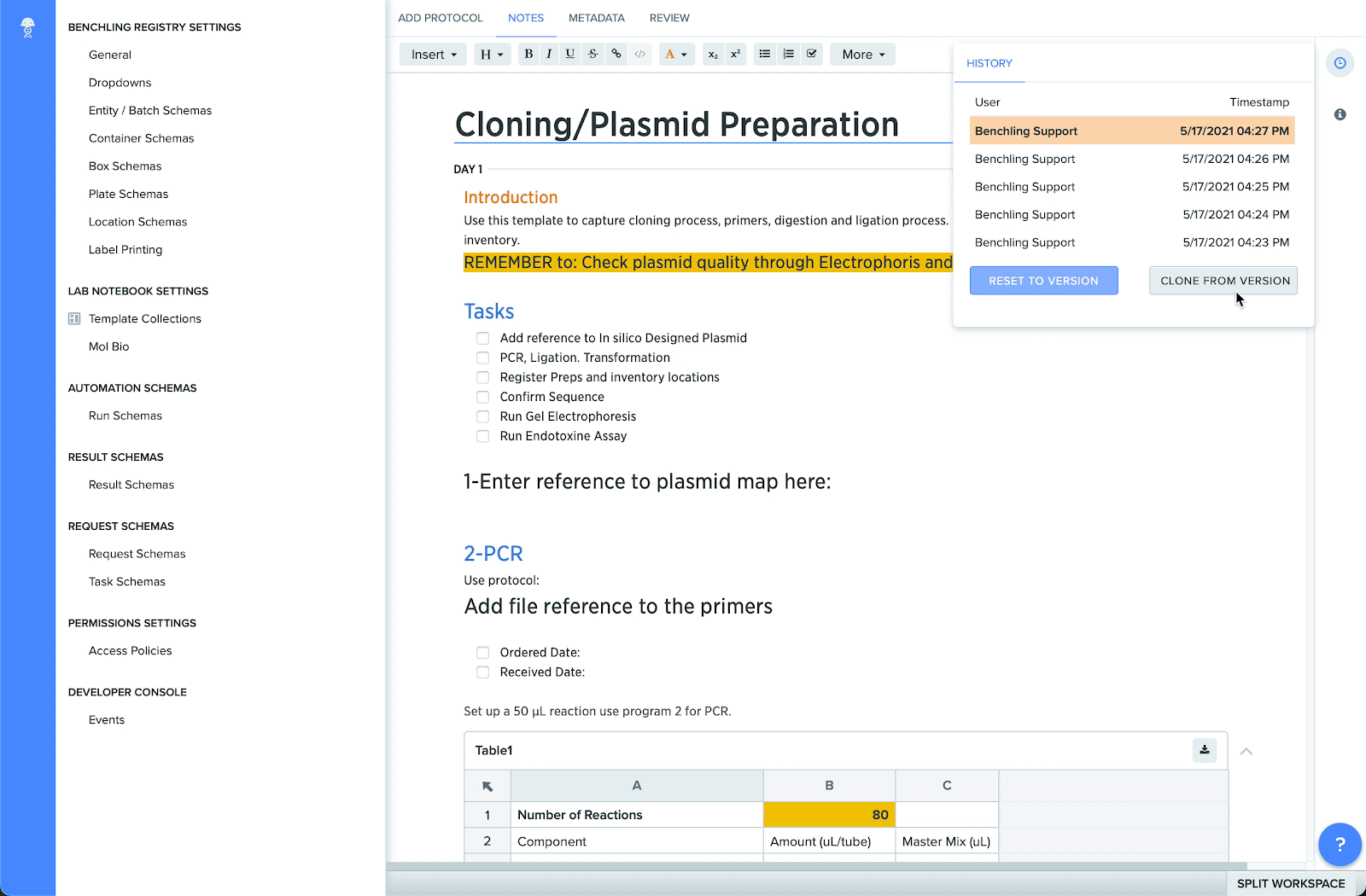
Support for column locking in tables
You can now lock columns in Notebook tables to preserve their contents and prevent other users from editing or deleting important information. This helps maintain the accuracy of data, while also making it simpler for users to fill out tables accurately. Locked columns will display a “locked” icon next to the column header.
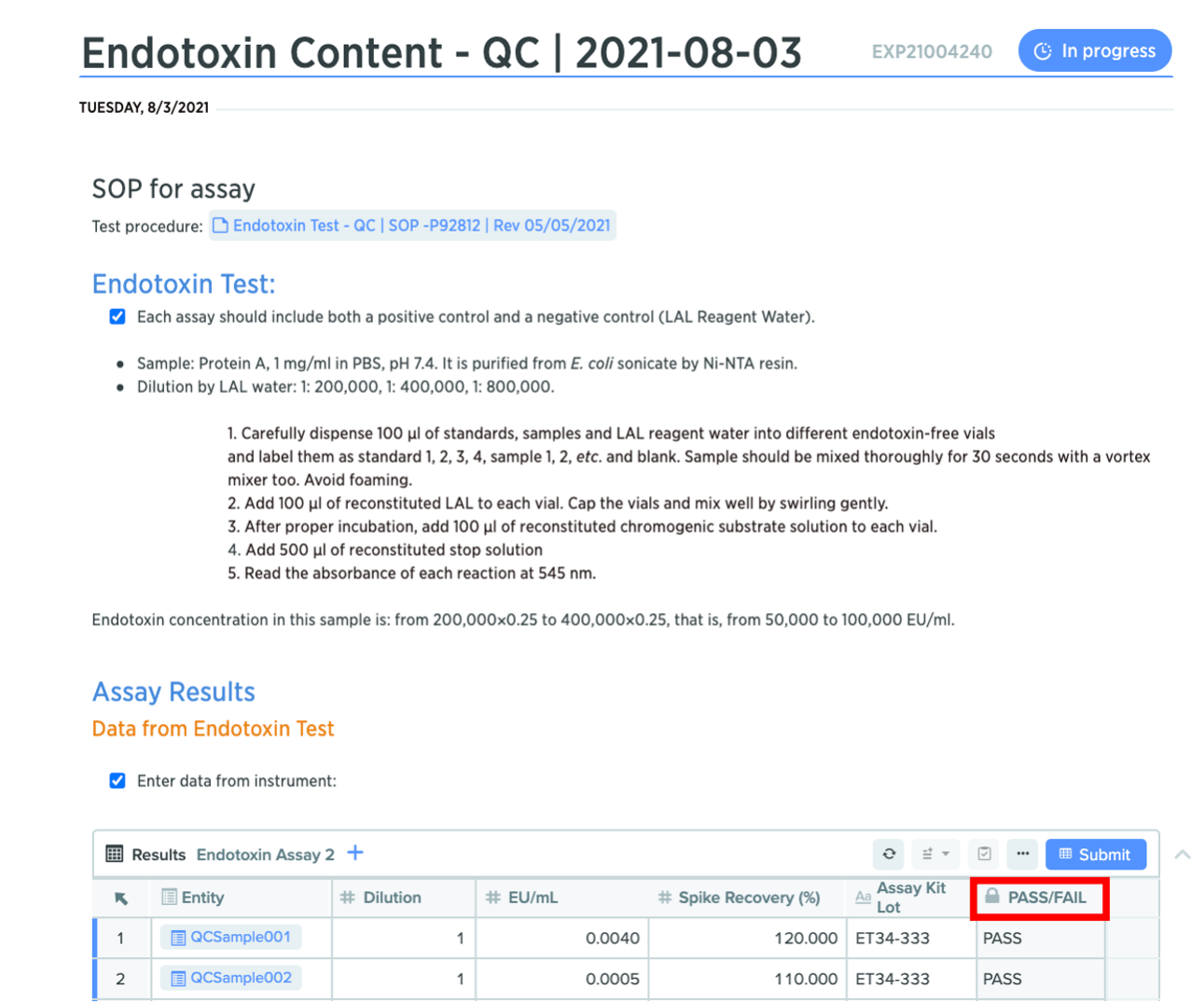
Improvements to table functionality and formulas
We’ve also added new formulas, and streamlined the user interface for Notebook entities and tables.
New formulas ISERROR() and ISBLANK(): Notebook tables now support ISERROR() and ISBLANK() formulas. The ISERROR() formula indicates when a cell contains an error, and the ISBLANK() formula indicates which cells are blank.
Notation for required fields: required fields in structured tables now display a red asterisk, indicating they’re mandatory to complete.
Table of contents: Notebook entries now contain a table of contents menu on the right side of the document, making it easier to navigate each entry.
Copy and paste tables: you can now copy and paste structured tables in Notebook templates.
Insights
With Insights, you can visualize, query, and share Benchling data to help generate actionable insights about your R&D pipeline, experiments, team productivity, and resource allocation. In this release, we’ve enhanced charts in Insights to include scatter plots with linear regression modeling.
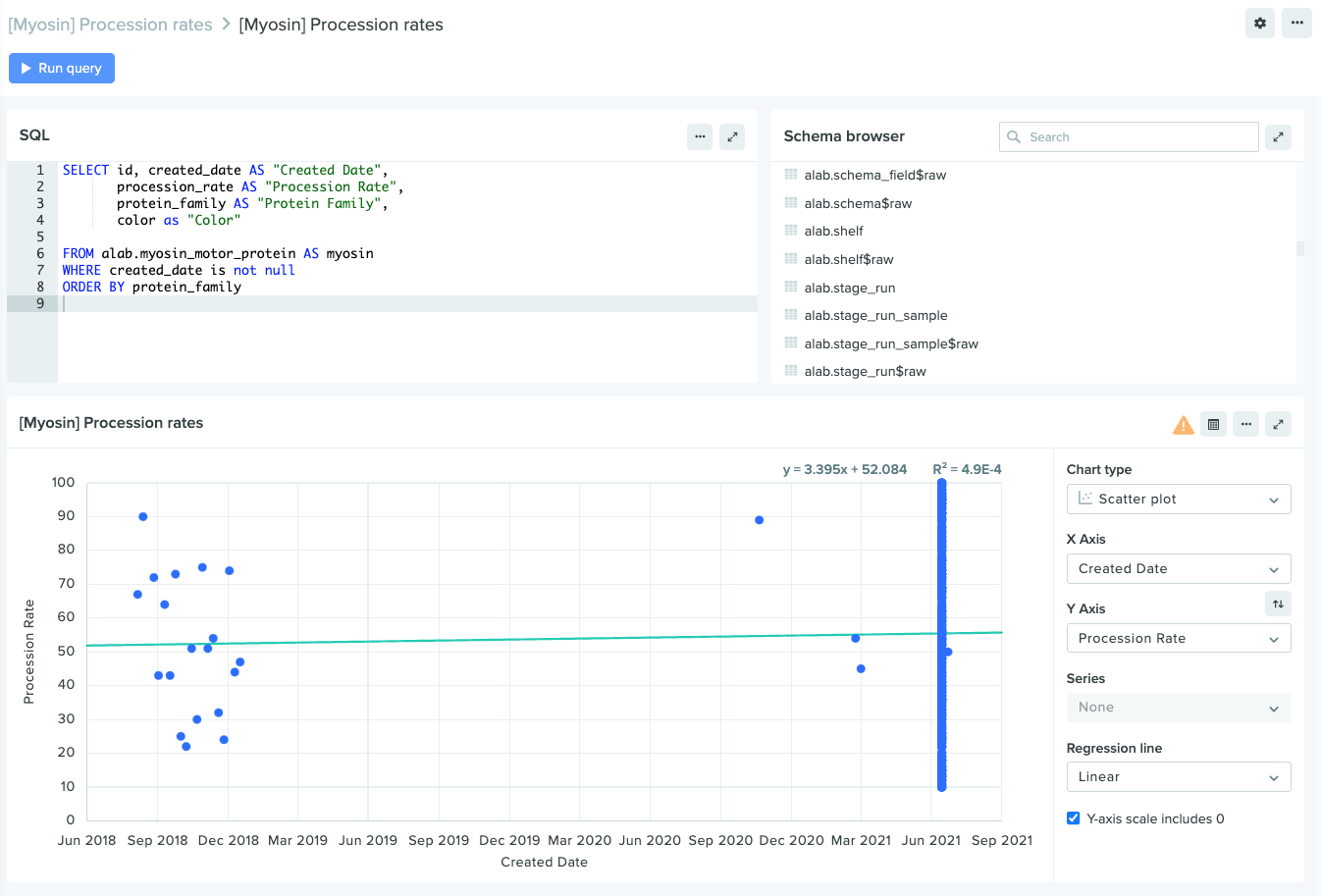
Support for linear regressions on scatter plots
Charts in Insights now provide a variety of helpful new visualization tools, including scatter plots and linear regressions. These new chart types make it easier than ever to model the relationship between two different variables — for example, the fluorescence of a myosin protein over a set time period — illuminating trends within your data to help answer key R&D questions.
Molecular Biology
Benchling Molecular Biology is an in-silico design tool that supports a wide array of molecular modeling workflows, such as primers, digests, and CRISPR guides. In this release, we’ve improved the Molecular Biology user interface to simplify and speed how teams edit, update, and annotate nucleotides, with an emphasis on enabling teams to complete actions in bulk.
Streamlined functionality for editing chemically modified oligos
Earlier this year, we announced new capabilities for adding chemical modifications to DNA and RNA oligos. In this release, we’ve improved the user experience and interface for editing these oligos.
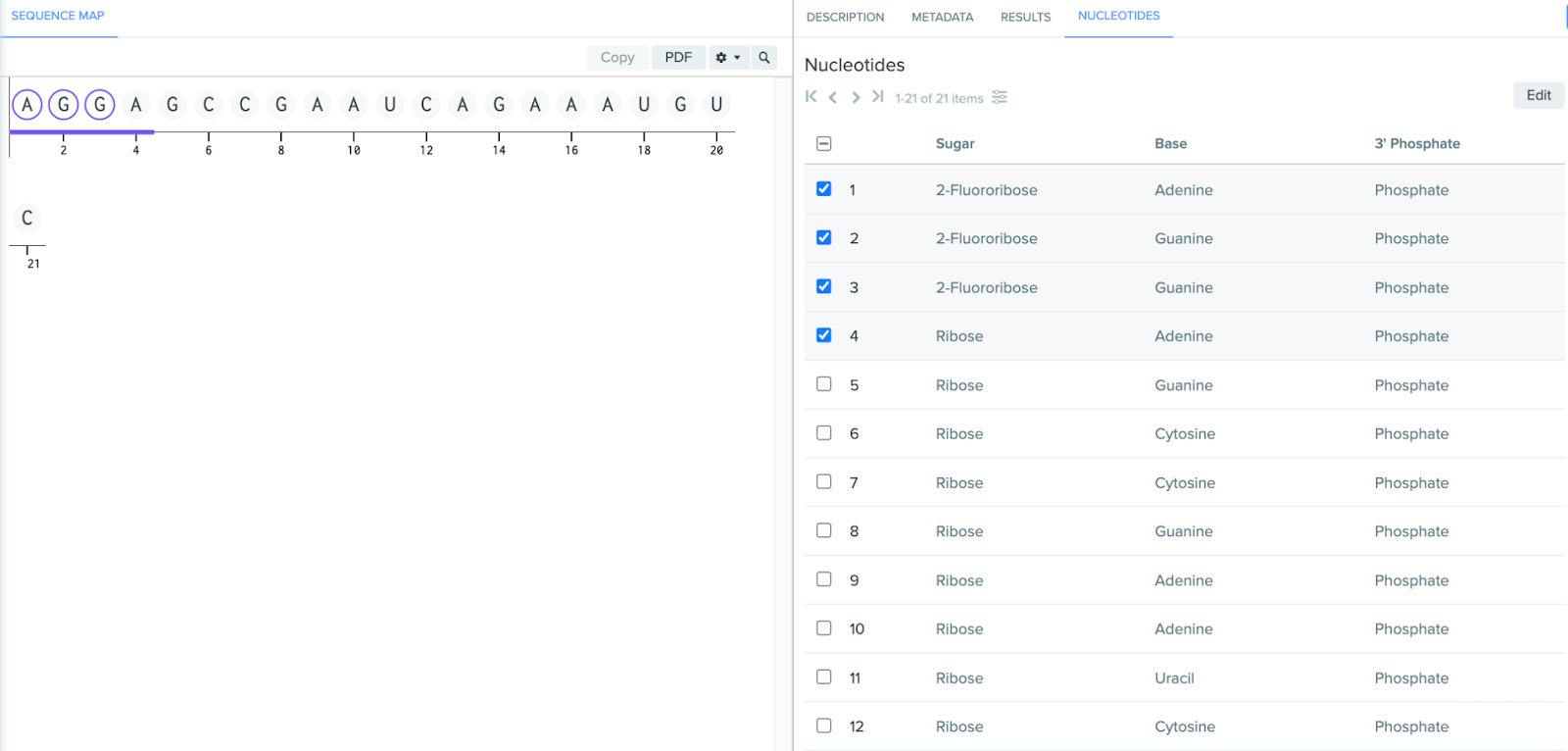
Visual cue when editing monomers: when selecting a monomer for editing, it will now appear in a light orange color within the sequence map and nucleotide list.
Bulk edit nucleotides: within the nucleotide tab, you can now select and edit multiple monomers at once, which makes it easier and faster to modify oligos.
Enhanced search functionality across oligos
You can now run exact- and substring-match searches across HELM representations of oligos, making it easier to search for these oligos with HELM.
Enhancements to DNA and amino acid (AA) sequences
We’ve added several new tools to streamline and simplify common tasks in Molecular Biology.
Bulk update AA residues: you can now update multiple AA residues at once when you update these entities via the CSV workflow.
‘Notes’ field in DNA sequences: the “annotations” menu within DNA sequences now contains a text box for notes.
Bulk remove annotations: from the expanded project menu, you can now bulk-remove annotations, attached primers and translations from a DNA sequence.
Registry and Inventory
Benchling Registry and Inventory work together to power your experiments through connected data, whether that’s data on sequences, samples, or experimental results. With Benchling Registry, you can standardize, connect, model, and track samples across all your experiments. Benchling Inventory provides a digital window into your physical lab, enabling you to track vials, wells, batches and more, while linking each of those elements to relevant results. This release includes streamlined schema naming and filtering, and clearer help text to guide you through the process of archiving samples.
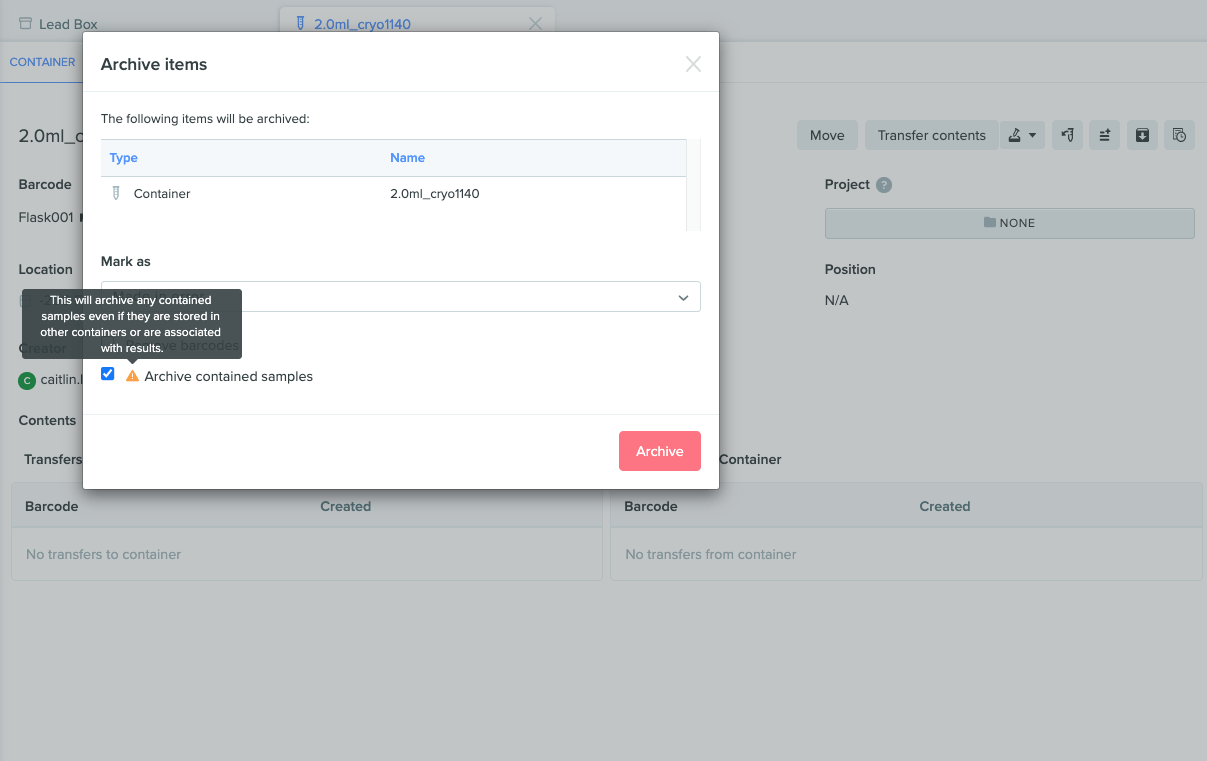
Improved warning text when archiving data
When archiving any sample within a container, Benchling now displays more specific warning text to clarify the impact of archiving this data. The text now states, “This will archive any contained samples even if they are stored in other containers or are associated with results.”
Improvements to the Registry and Inventory user interface
We’ve also updated the user interface to accelerate how you complete common tasks, specifically naming templates and filtering schemas.
Drag-and-drop naming templates: you can now drag-and-drop and reorder components in a template name to easily modify it.
Automatic schema filtering: when typing filter criteria into the admin page, schema results will refine in real time based on the text entered.
Lab Automation
Lab Automation enables you to integrate lab instruments, like liquid handlers and plate readers, into Benchling to scale experiments, increase scientist productivity, and sync research across software and hardware. In this release, we’ve added new parameters for configuring runs and refining processing steps.
Improvements to run configuration
We’ve streamlined the “run config” user interface, and made it easier to maintain data integrity between steps.
Preserve relationships in lookup config: when configuring lookup steps during a run, you now have the option to maintain per-row relationships of data based on the previous lookup step. Runs now contain a box labeled “Do not preserve relationships between lookup steps.” Leaving this box unchecked will preserve information between steps.
Hide archived fields: during runs, you can now hide archived fields for a simpler view of the relevant run components.
Default values: you can now set default run values within entry templates.
Simultaneously process samples and results: when processing samples, you can now add a step to the “transfer bulk” adapters to process results as well.
New processing steps
This release includes three new processing steps for tailoring more precise runs.
MELT processing step: you can now unpivot or flatten data in tables to consolidate it into two columns, variable and value.
ARITHMETIC processing step: you can now use arithmetic (+, -, *, /) to perform functions between two columns.
PLATE_SHAPE error message: if a plate schema is larger than the corresponding CSV, the plate shape processing step will now display an error message.
Platform
The Benchling platform connects capabilities and data across all Benchling tools, in order to provide a simple, unified user experience. In this release, we’ve expanded access controls within the platform, and added new APIs for more precise filtering, archiving, and capture of results.
Tighter control over user access to developer capabilities
Within the tenant admin console, you can now manage user access to the developer platform. This feature builds on existing capability management functionality, which enables admins to provision or remove user access to apps within Benchling — providing granular control that helps ensure only authorized users, teams, and orgs have access to relevant apps.
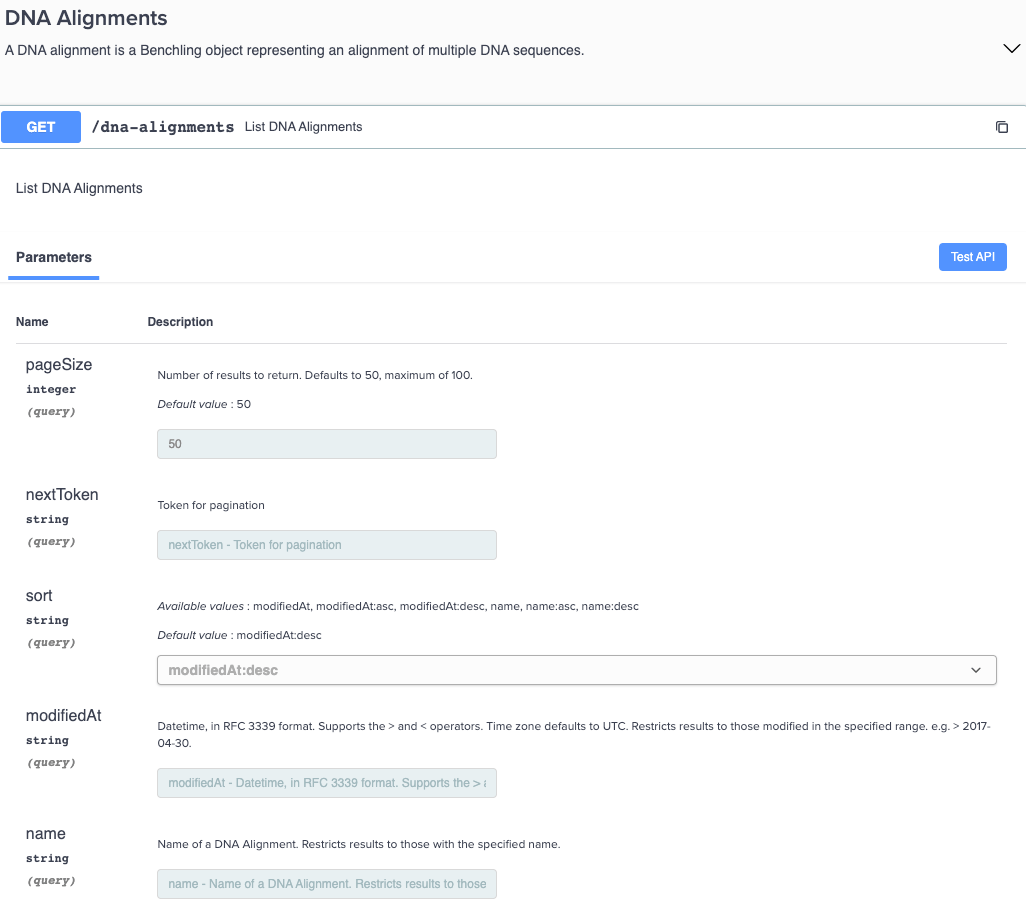
New GET endpoints for DNA alignments
You can now retrieve a list of DNA alignments via a new GET endpoint in the API. This endpoint includes the Benchling web user interface, URL, and information on when the alignment was modified and created.
API updates
Benchling APIs are built to match the speed and agility of modern R&D, making it easy to extend Benchling into your broader digital ecosystem. We regularly enhance and add new APIs to further increase Benchling’s flexibility and expand its range of potential integrations.
Support for solid units: container APIs now support solids in addition to liquids, fulfilling a wider variety of scientific use cases. To facilitate this change, the default unit of measurement for container APIs has been changed from “volume” to “quantity.”
Filter APIs by creatorID: you can now filter API search results by user, or by a set of users.
More granular archive filtering for list endpoints: the archiveReason query (part of every endpoint) provides more detailed options for defining archive parameters. You can now select one of four archive parameters from the dropdown list, including: “only include archived items,” “include items archived for a specific reason,” “include items archived for any reason,” and “include both archived and unarchived options.”
Table types added to “entries” API: the “entries” API now contains information on the position, name, and schema for the following tables: results tables, registration tables, plate creation tables, box creation tables, and mixture prep tables.
Documentation for long-running tasks: we’ve created new API documentation for long-running tasks. You can access this detailed documentation here.
Ability to archive dropdowns: you can now archive options within a dropdown menu via the API.
API parity for POSTing and PATCHing schema fields: the APIs for POSTing run schema fields, PATCHing run schema fields, and POSTing result schema fields are now more consistent with other schema APIs.
Powering breakthroughs for over 1,300 biotechnology companies, from startups to Fortune 500s
