Benchling Product Release Newsletter: September 2019
September's Updates: Registry Labels, Plate Creation, CRISPR Guide Design, Request Samples
Welcome to the September edition of our monthly product release newsletter! In this month's update, we will cover major new features in Benchling like registry labels, the new plate creation table for Notebook, improvements to CRISPR guide design, and our API that allows for Request updates.
Registry labeling
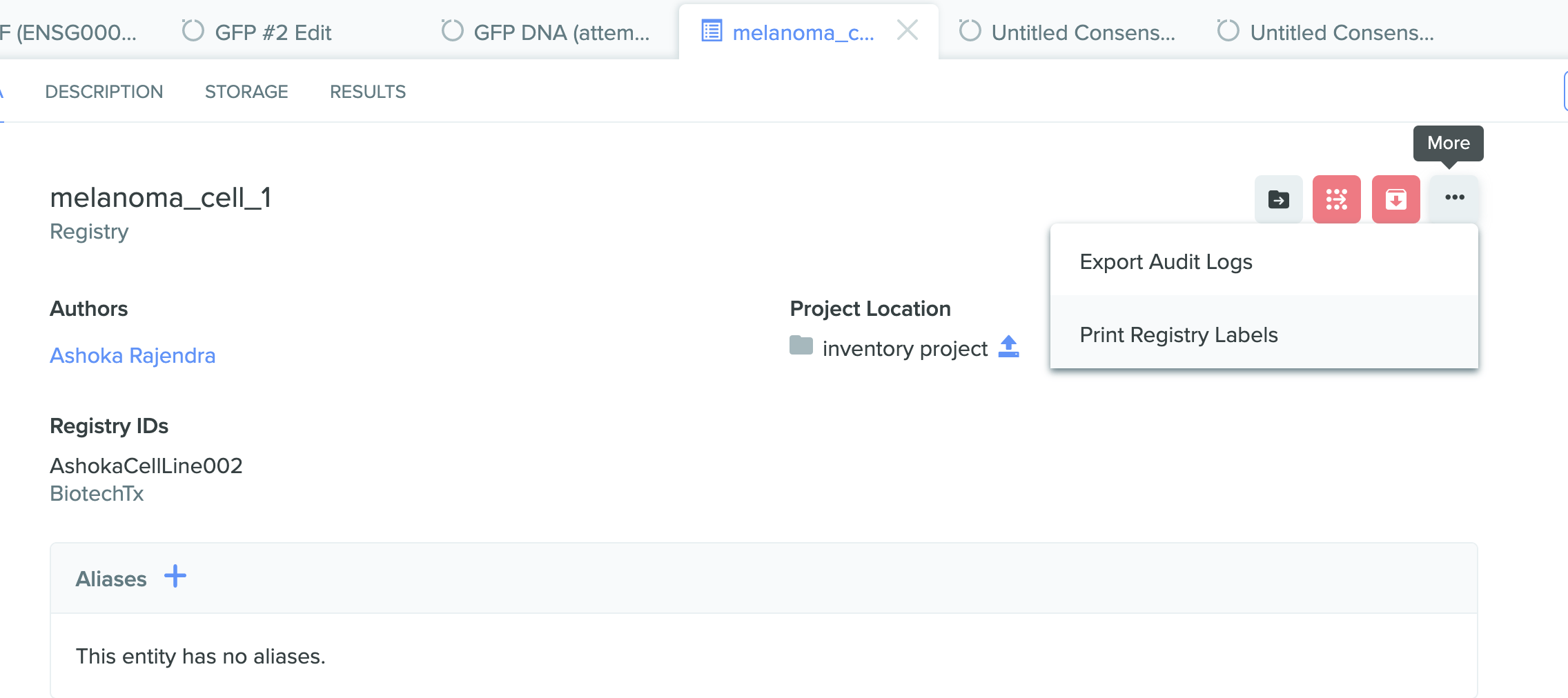
Creating registry labels for any registered entity is now enabled in Benchling Registry. Be it equipment, filters, raw materials, solution recipes, packed columns, resins, pools, or virus stock, customized registry labels can now be created for any entity that has a Registry ID. This feature is also integrated into Benchling Notebook allowing you to scan Registry labels into tables in notebook entries directly to indicate use in a given experiment.
1) Printing labels from Registry entry
2) Printing labels from Registry search
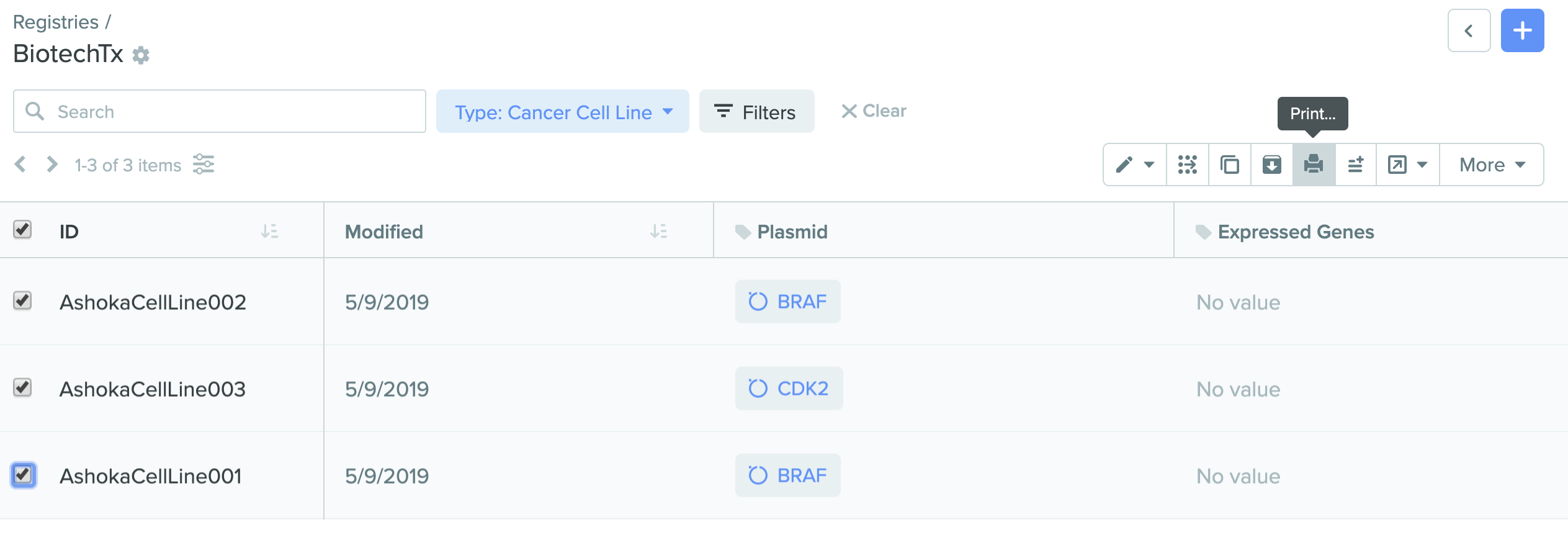
Plate creation table in Notebook
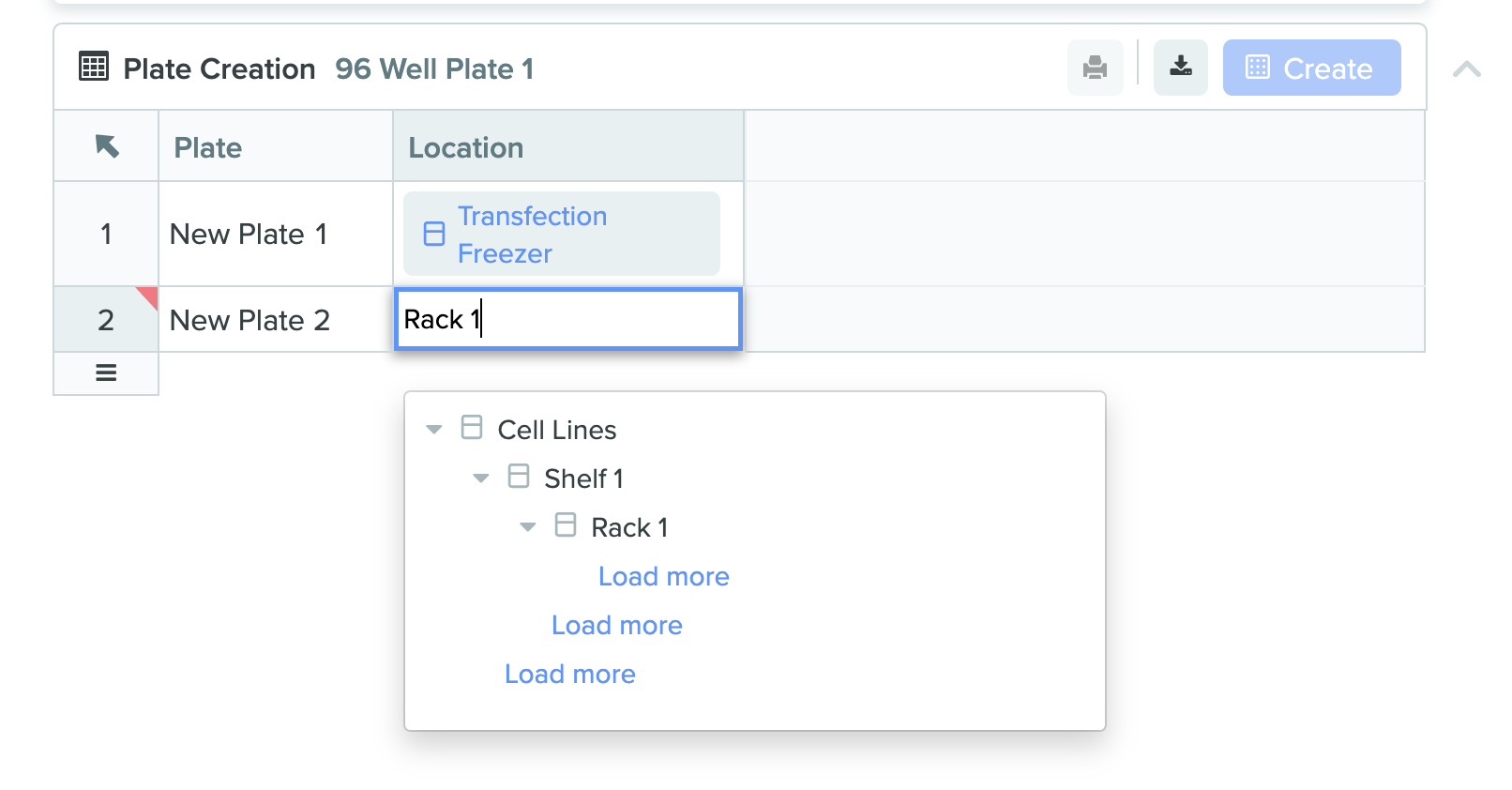
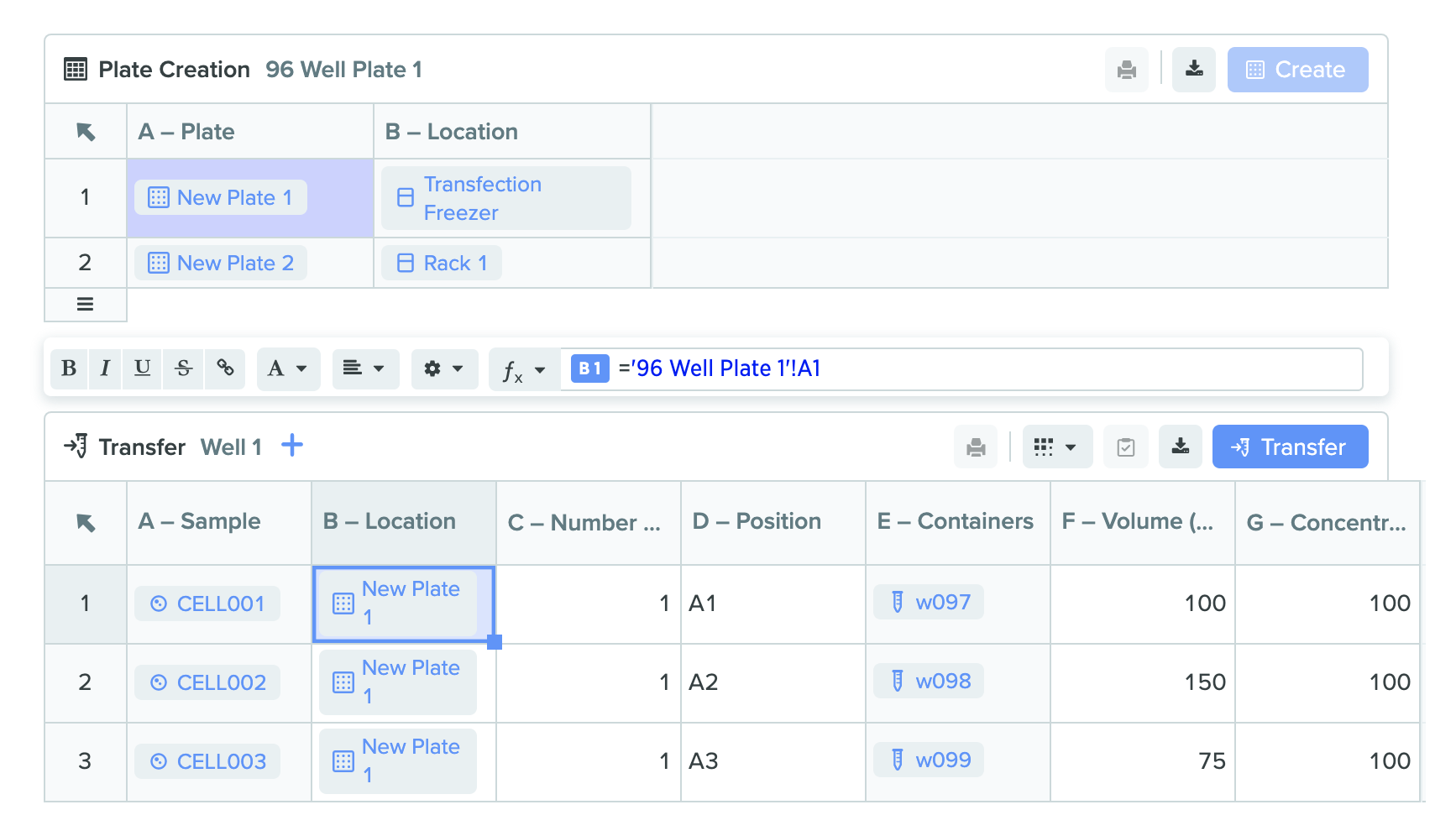
We added a brand new structured table type in Notebook entries called Plate Creation Table. This feature provides an easy to use UI to create multiple plates, scan existing plate barcodes into the table, navigate location hierarchies through an improved location selector, and set-up more complex plate creation templates to ensure standardization and compliance. Plate creation table works in tandem with transfer table in a Notebook entry to streamline assay preparation from sample creation to sample use. Additionally, this new table type can be leveraged for lab automation applications such as creating many destination plates to use in a liquid handling run. Example screenshots of the new feature are shown below:
1) Plate creation table with new location selector
2) Plate creation table with formula from transfer table
CRISPR guide design improvements
The CRISPR tool on Benchling received an update directed towards providing more flexibility to our users. Below are some of the key highlights:
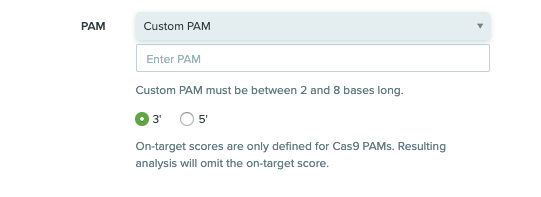
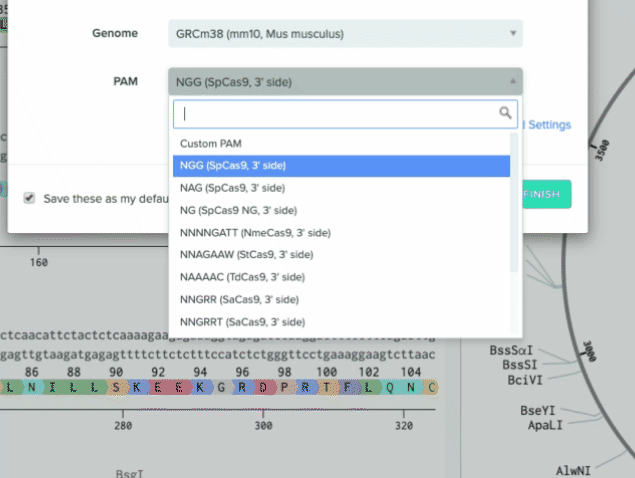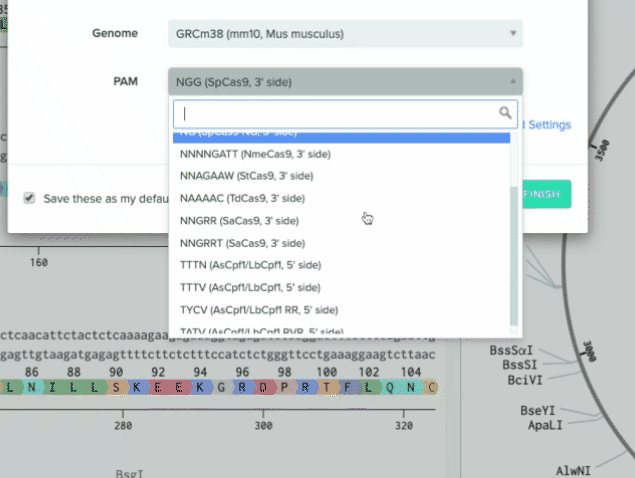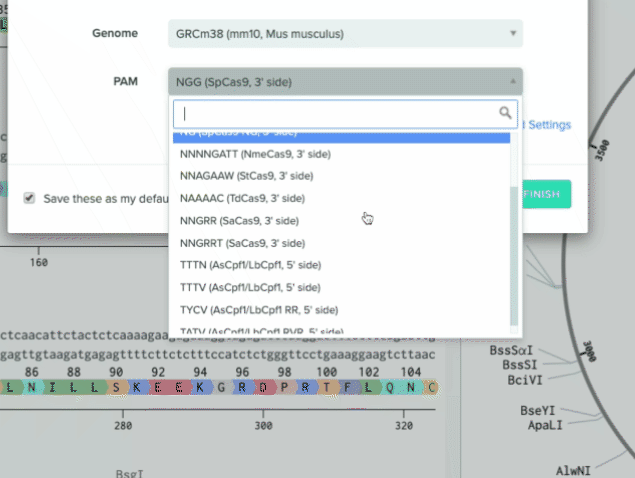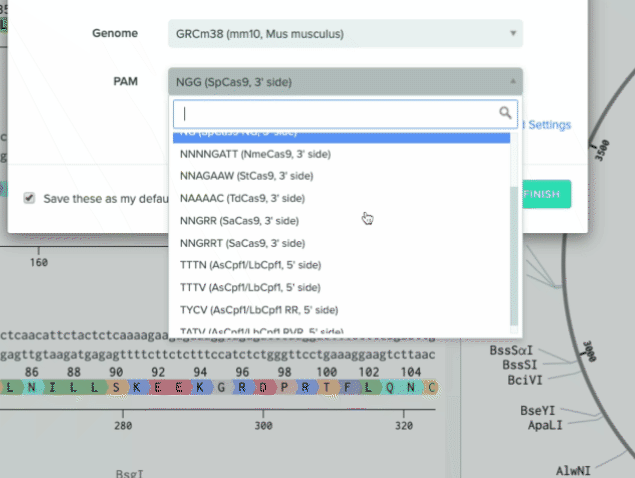
Benchling can now effectively support any CRISPR nuclease and nuclease variant with the ability to create a custom PAM site on the 3’ or 5’ side of the guide RNA
Option to toggle on/off spCas9-specific positional weights for off-target scoring to better fit your research needs
We also added four additional nucleases to our default list to support a wider range of use cases
RVR variant of As/Lb Cpf1. PAM site = TATV
RR variant of As/Lb Cpf1. PAM site = TYCV
2 additional options for SpCas9 that target NG and NGA PAM sites
1) Creating a custom PAM
2) Additional nucleases added to our default list
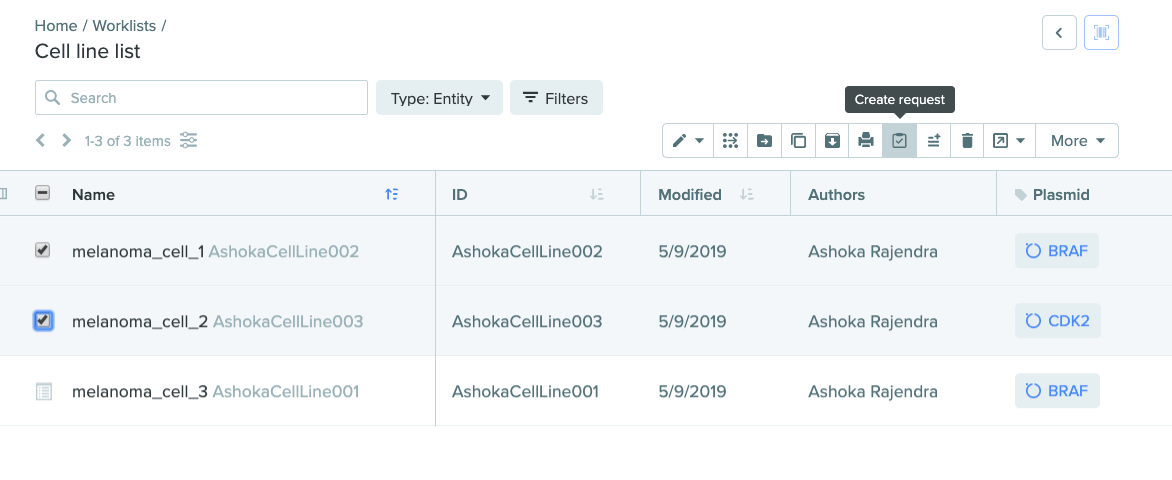
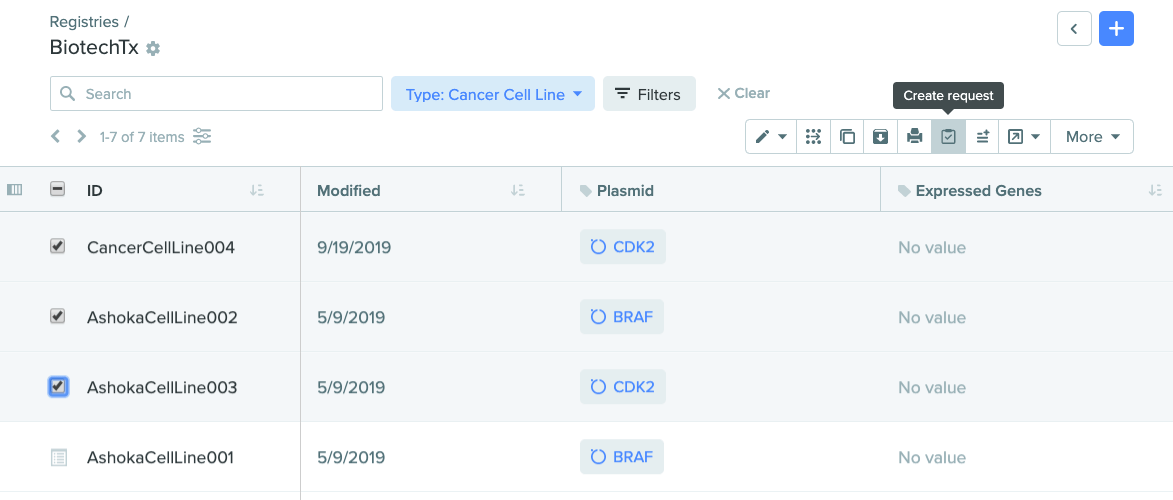
Create requests with entities from toolbar
Creating a request with entities is now easier than ever. Requests are often made with multiple entities at a time. In addition to the ability to create a request with entities directly from a registration table, you can now create a request in the following new ways:
Gather entities into a worklist and create a request directly.
Generate requests directly from registry search
Create requests from the expanded project list view
This update provides convenient ways to create requests with multiple entities outside of a notebook entry. It also unlocks the power of the search function to find entities and create relevant requests quickly. Note, that this feature supports only entities. Containers and batches are not supported at this time.
1) Create request from worklist
2) Create request from registry search
Update Request samples in the API
We released a new API that allows users to update requests directly through the API. Specific functionality now enabled includes:
Create a request with samples organized into sample groups in the API
Add, update, or remove samples or sample groups on existing requests in the API
Execute sample groups without tasks in the API
This new API better supports lab automation use cases where instruments are updating requests or fulfilling requests directly on Benchling. Additionally, custom business logic such as executing requests on samples different that those originally requested or the addition of samples to the original request are now better supported.