Unlocking High Throughput Transcription Factor Mapping With Biotin-DAP-seq
Guest author Leo Baumgart is a scientist in the Sequencing Technologies group at the Joint Genome Institute, where he works on developing new methods for studying genetic control elements in plants and microbes.
At Berkeley Lab, we’ve developed a pair of new methods that we’re calling biotin-DAP-seq and multiDAP. These methods can be applied to map transcription factors (TFs) to their target genes 40x faster and 10x less expensively than previous methods. For the first time, the time and monetary costs are low enough to do high-throughput pull down for thousands of samples. With this technology, labs and biotech companies can:
Translate DNA sequences into affinity labeled proteins
Isolate proteins for downstream characterization
Apply to transcription factor (TF) binding site discovery methods (e.g., DAP-seq, SELEX), RNA protein-binding site discovery, and protein-protein interaction studies
Mapping transcription factors (TFs) to their target genes is fundamental to illuminating how gene networks function. However, information regarding TF-gene interactions is lacking for most organisms. Previously, the primary technical bottleneck to acquire such information was the significant upfront investment required to clone and express the tagged TFs used in in vitro assays.
We’ve developed Biotin-DAP-seq, a streamlined clone-free workflow where tagged TF proteins are expressed from DNA templates that are polymerase chain reaction (PCR)-amplified directly from genomic DNA, cDNA, or plasmids.
We depend on Benchling for tracking user samples through the entire process, from the time they arrive in the mail until they are ready to load on the sequencer. By modeling each step of the DAP-seq workflow in Benchling and integrating our liquid handler methods we can ensure that we keep a detailed record and verify sample barcodes at each step.
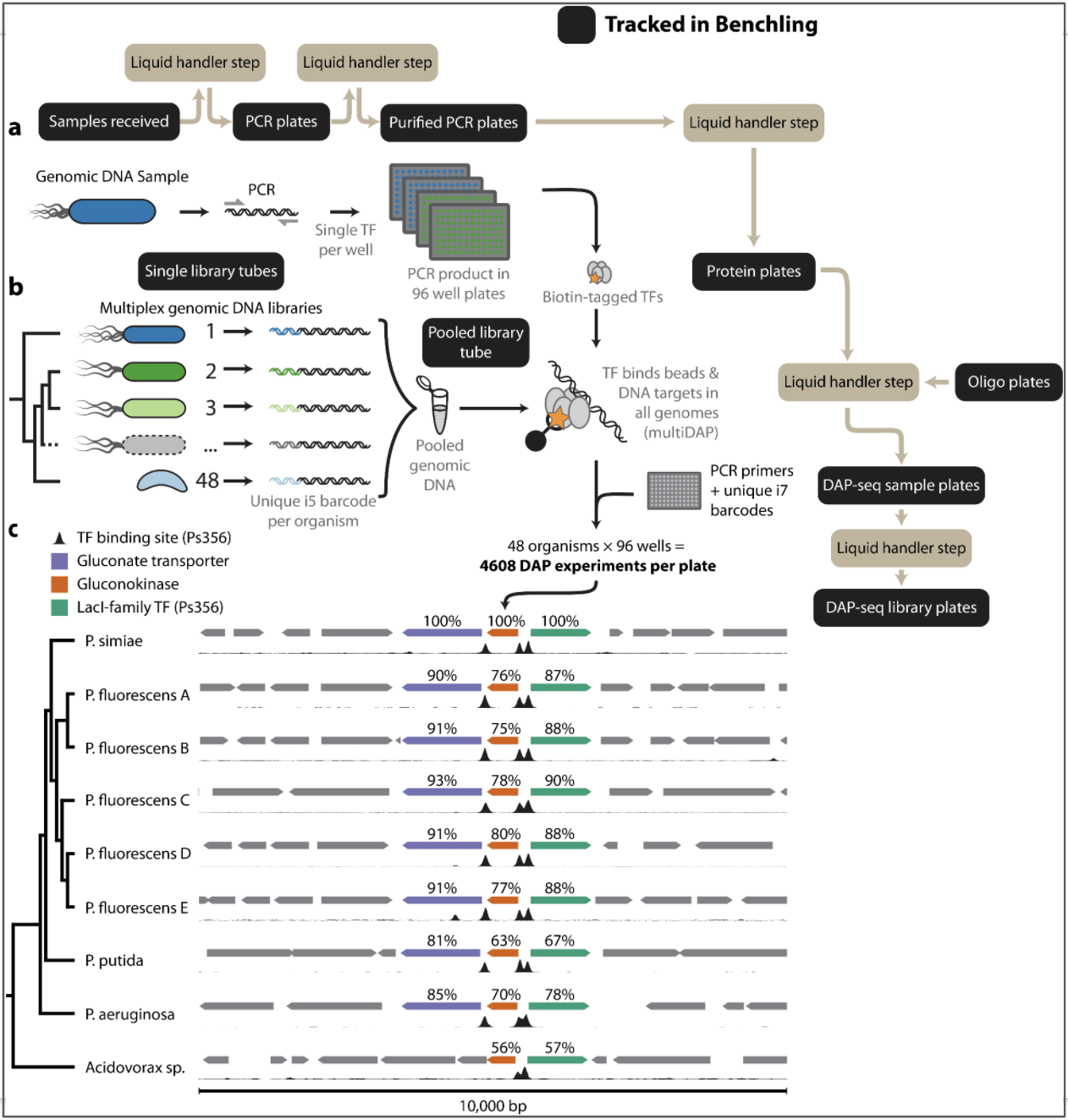
Ultimately, biotin-DAP-seq removes the need for cloning and purifying TFs and therefore enables applications at scale to any organism of interest. It is applicable to TFs from all domains of life including bacteria, archaea, and eukaryotes. Surveying TFs across all kingdoms of life reveals themes of ancient conservation and rapid evolution of regulatory modules, deepening our understanding of the genetic underpinnings of organisms. While much of our existing knowledge has been limited to studying a limited set of model species, biotin-DAP-seq and multiDAP will enable rapid characterization of TF-gene interactions in diverse non-model organisms. This information will lay the groundwork for new synthetic biology applications and engineering plants and microbes.
If you'd like to learn more about these methods, reach out to Leo Baumgart.
Contact us if you're conducting research with Benchling that you'd like to feature on the Benchling Blog.
Powering breakthroughs for over 1,300 biotechnology companies, from startups to Fortune 500s
