Introducing Protein Alignments on Benchling
With our new Protein Alignment feature and newly updated protein tools, you can import, align, analyze, and export your protein sequences right in Benchling.
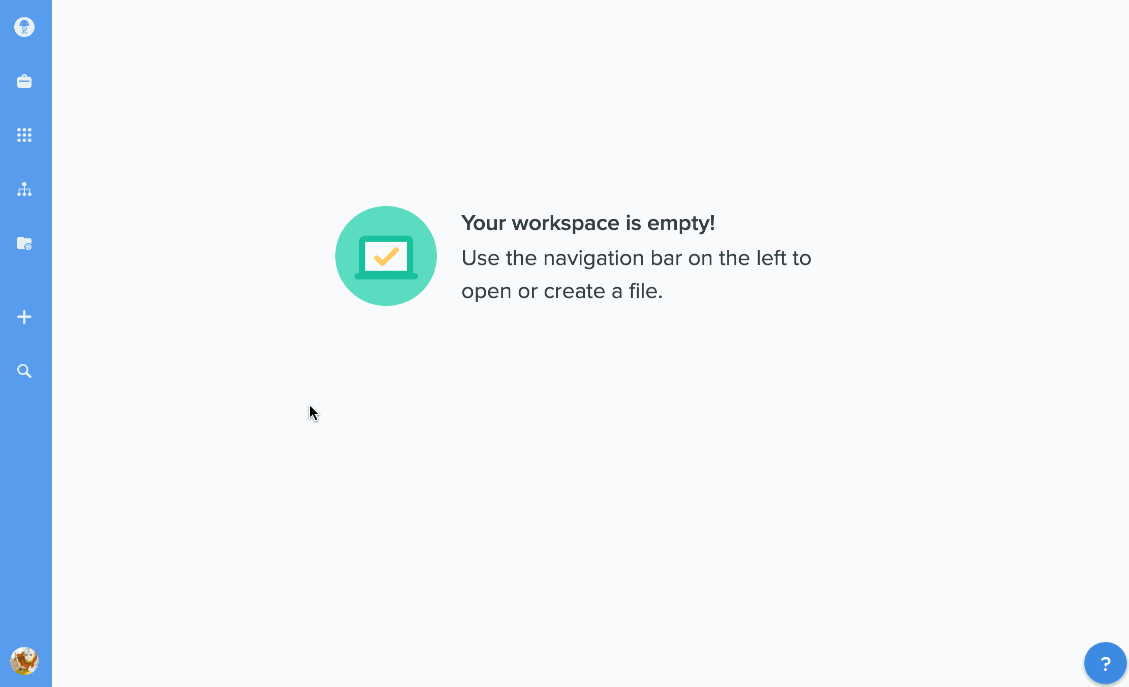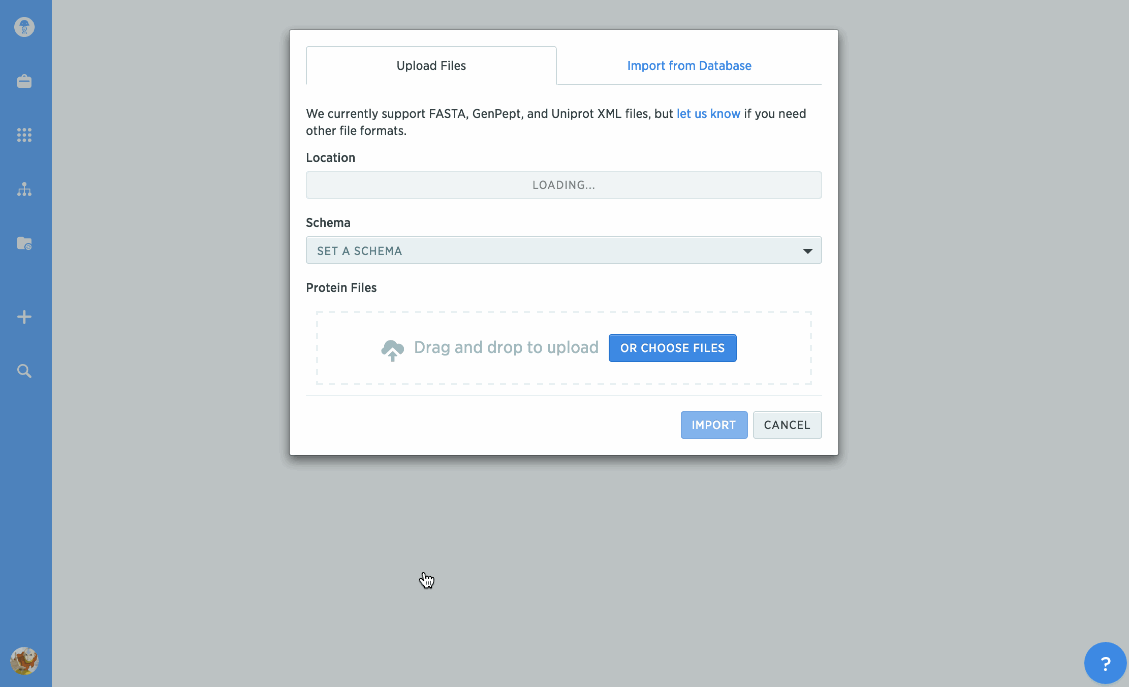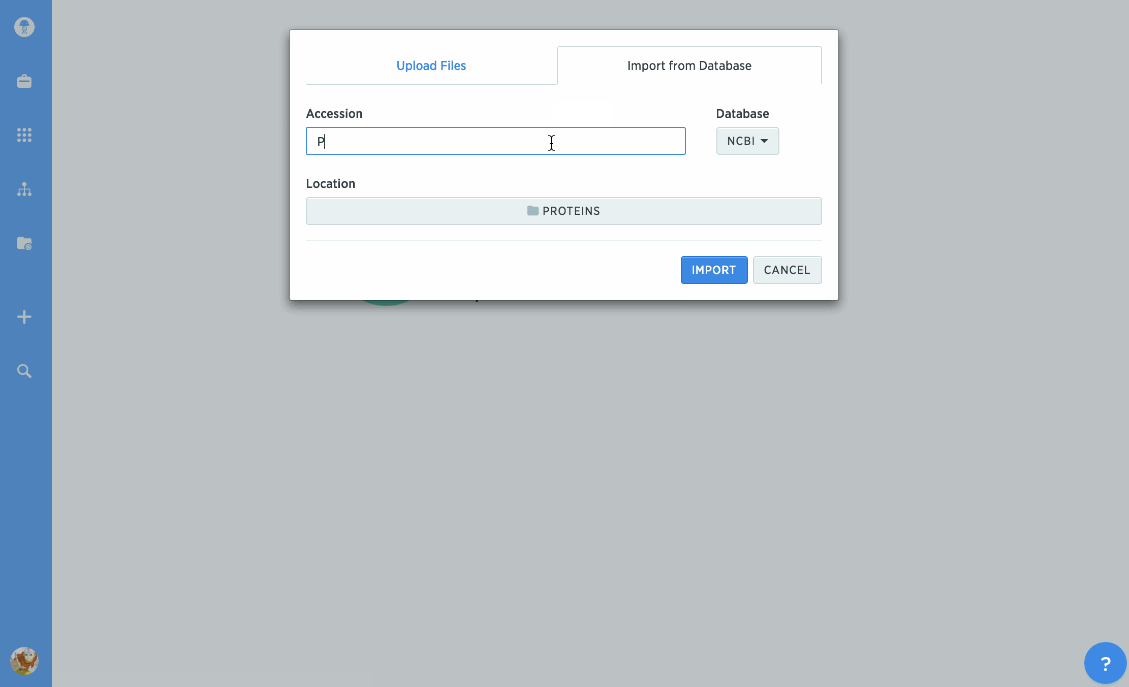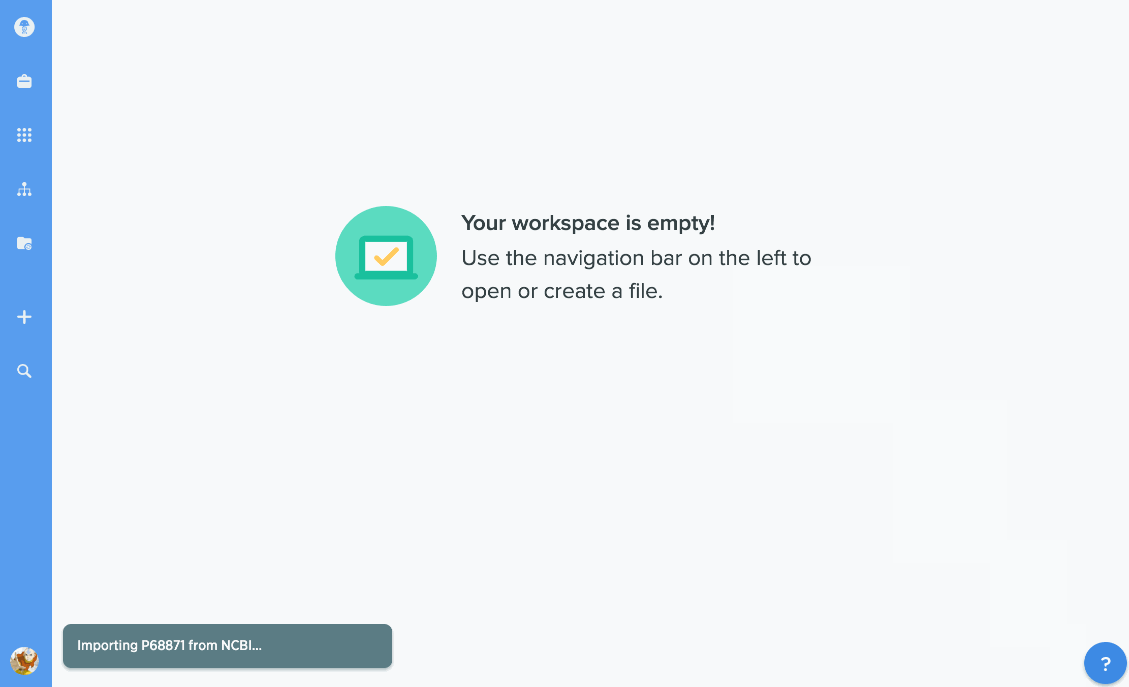
Import proteins from NCBI and UniProt
We've given our protein import tools an upgrade, so now you can import directly from databases like NCBI or UniProt. Just type in the accession number you're looking for to import it directly into Benchling. You can also import GenPept or UniProt XML files to keep track of annotations.
Perform consensus or template alignments
You can use our protein alignment tool to view consensus proteins, or compare proteins against a template. Alignments are powered by Clustal Omega and can support dozens to hundreds of proteins.
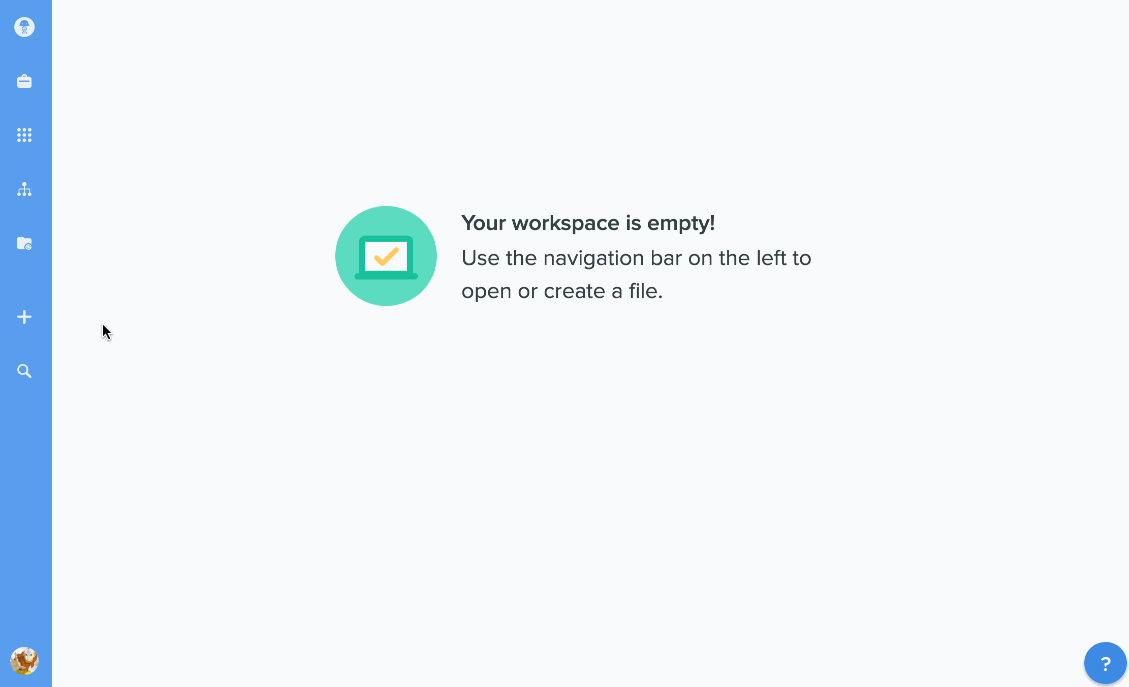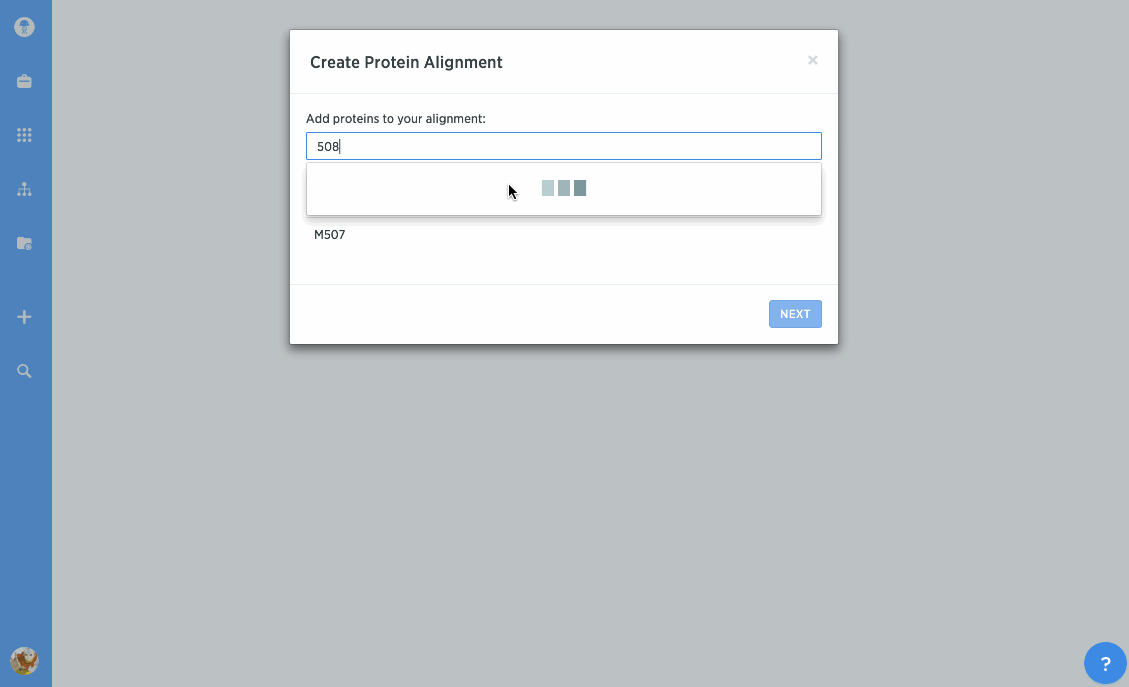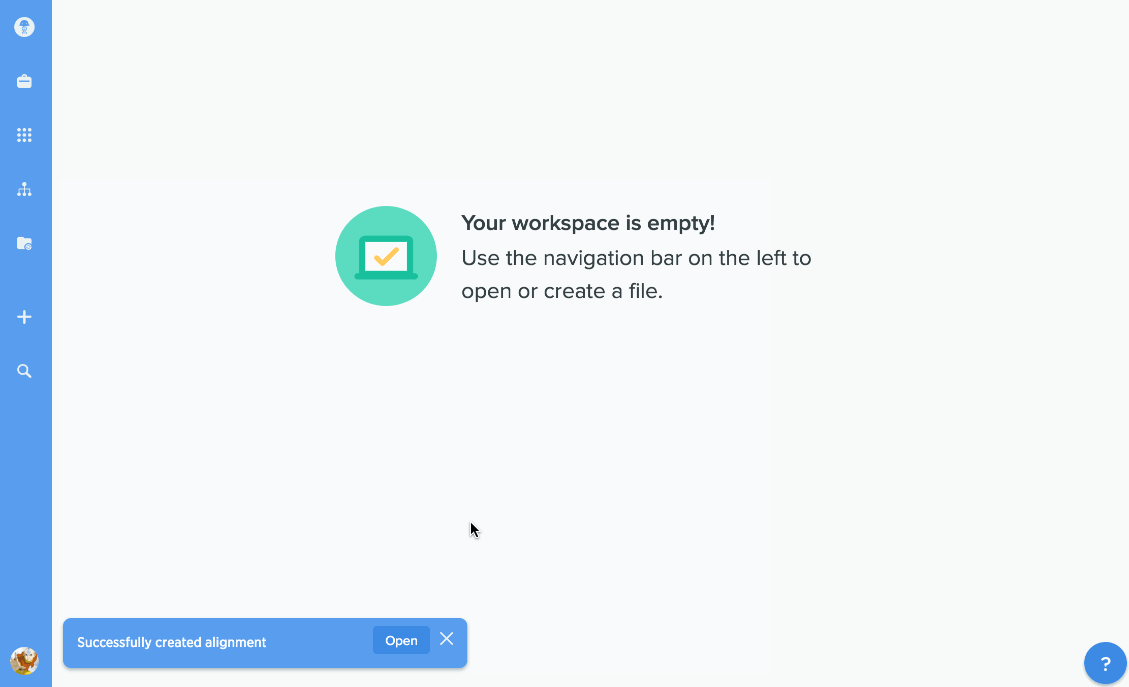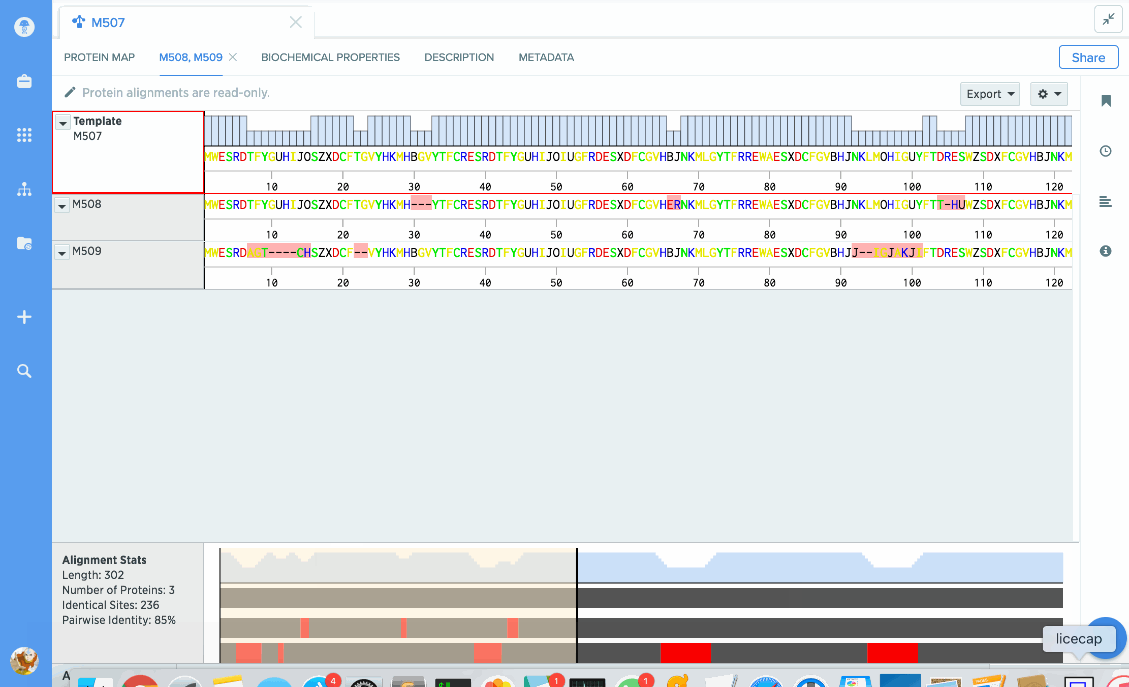
Analyze stats like % identity or hydrophobicity.
Our flexible visualizations provide you with exactly the information you need. You can see information about your current selection like length, identical sites, and pairwise % identity.
You can turn on graphs like hydrophobicity, pI, and % identity. A global summary map is also shown to give you an at-a-glance view of your alignment.
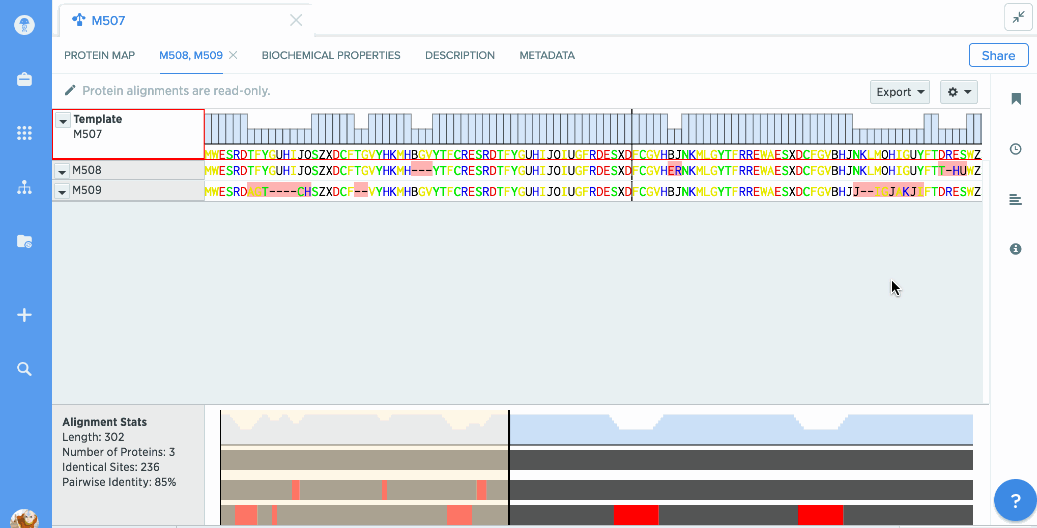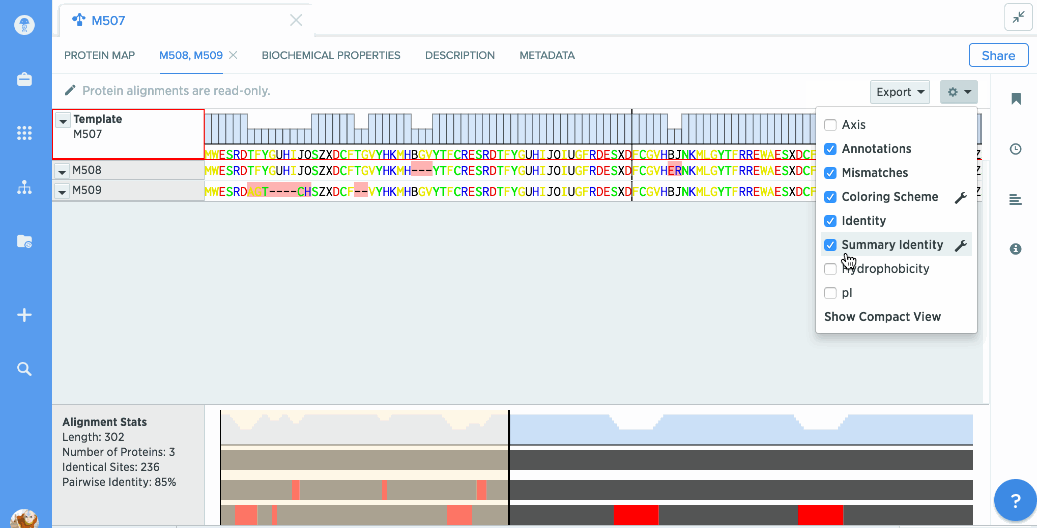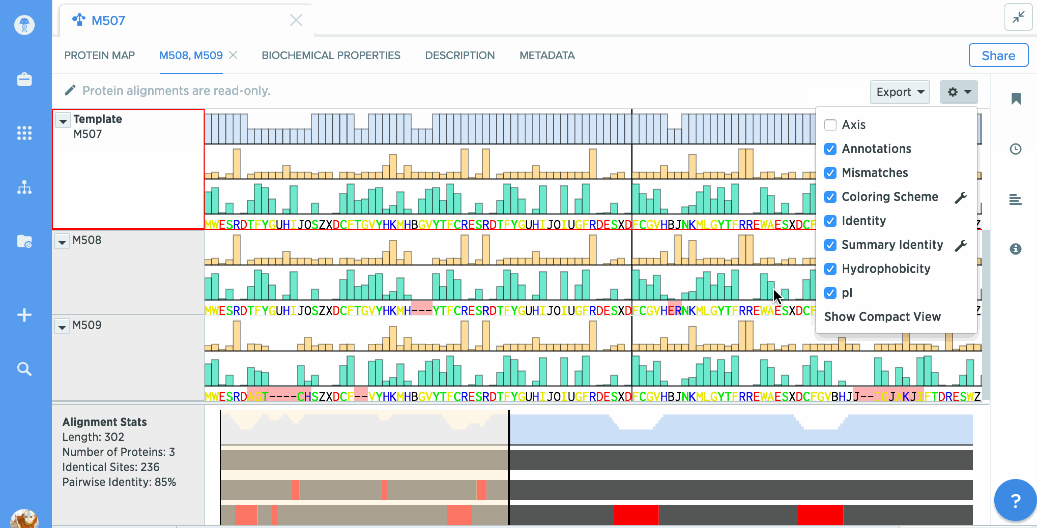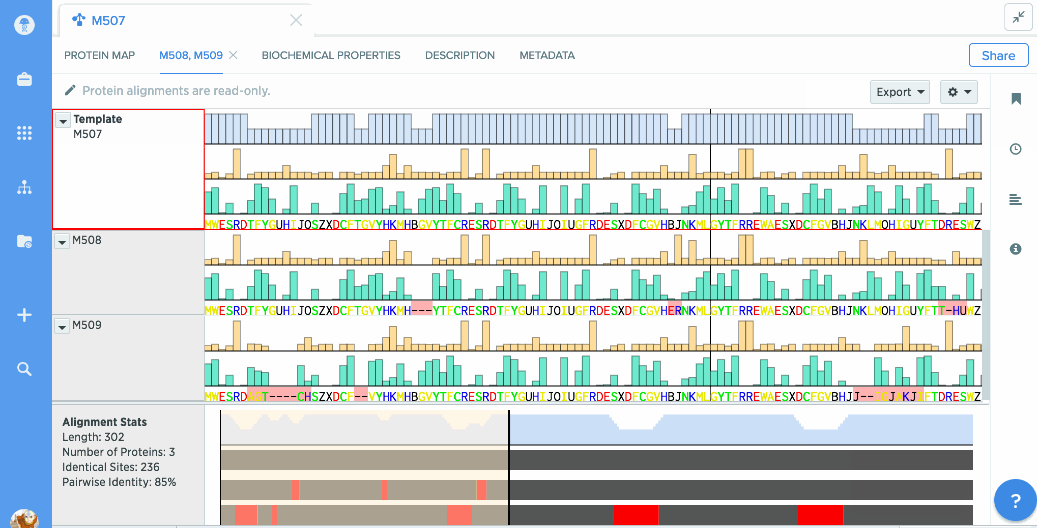
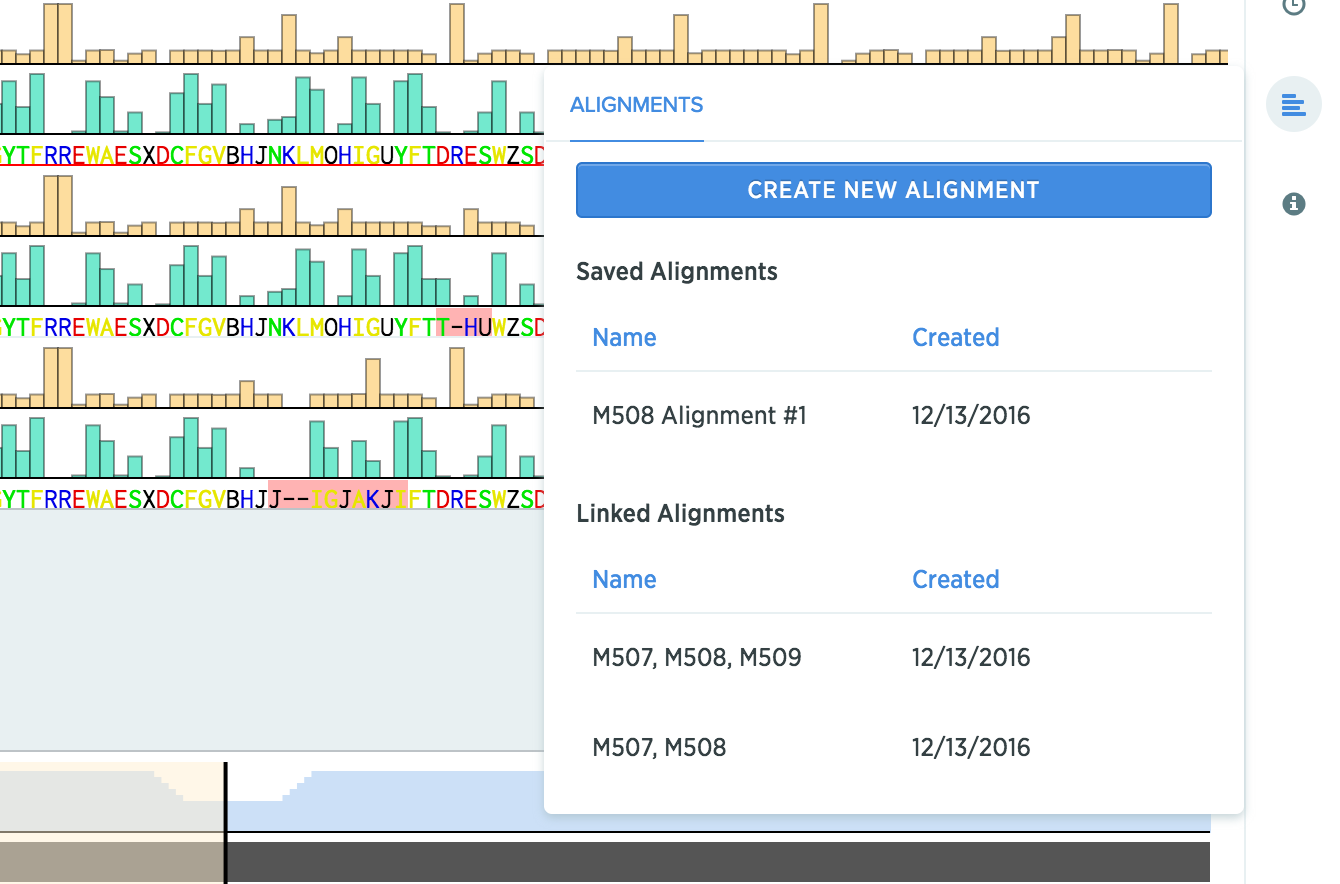
View alignments using a protein in one place
In the alignments panel, you'll be able to see any alignment that you've created using this protein, even if they live on a different protein.
Export as an image
Instead of trying to fit everything into a screenshot, use our easy-to-use image export to export your alignment as a .png and insert it directly into your presentations.
As always, try it out and reach out to us with any feedback!
Powering breakthroughs for over 1,300 biotechnology companies, from startups to Fortune 500s