Model siRNA and oligonucleotide conjugates consistently with Benchling
Working with hybrid modalities just got easier
Hybrid modalities, especially RNA conjugates, continue to enable scientists to pursue undruggable targets. Building on Benchling's previous releases for macromolecules at the intersection of chemistry and biology, comes the first in a series of updates allowing you to model oligonucleotide-based entities that have been chemically conjugated with small molecules. This allows teams to explicitly designate the chemical connections to improve the accuracy of tracking siRNA and ASOs in Benchling.
RNA therapeutics continue to draw research interest
RNA therapeutics are attracting growing research interest with over 1,600 drug candidates in preclinical and clinical development (1). Small interfering RNA (siRNA) and antisense oligonucleotides (ASOs) are the two leading RNA therapeutics platforms with the highest number of approved drugs on the market (2-5). Chemical conjugation of these RNA therapeutics has emerged as a major approach to the successful delivery of these drugs (2-7). For example, GalNAc (N-acetylgalactosamine) conjugation has shown to be effective in highly specific delivery of siRNA to hepatocytes (2-6).
Modeling siRNA and oligonucleotide conjugates can be cumbersome
Visualizing siRNA and antisense oligonucleotides (ASOs) with chemical modifications and conjugations is a challenge. The hierarchical editing language for macromolecules (HELM) developed by the Pistoia Alliance helps to standardize the molecular line notation of siRNA and ASO conjugates (8). However, the actual design experience of creating and editing these molecules is highly variable depending on the tool or software vendor used.
The key challenge is in the navigation between the complex polymer, simple polymer (or sequence), monomer, and atomic view of the siRNA and ASO conjugate structure. Scientists need to model siRNA and ASO conjugates with fidelity across the entire hierarchy from the composite polymer level to the atomic level. This helps scientists validate connections, confirm scientific accuracy, register conjugated sequences, and calculate molecular properties.
Visualize and design siRNA and oligonucleotide conjugates with Benchling
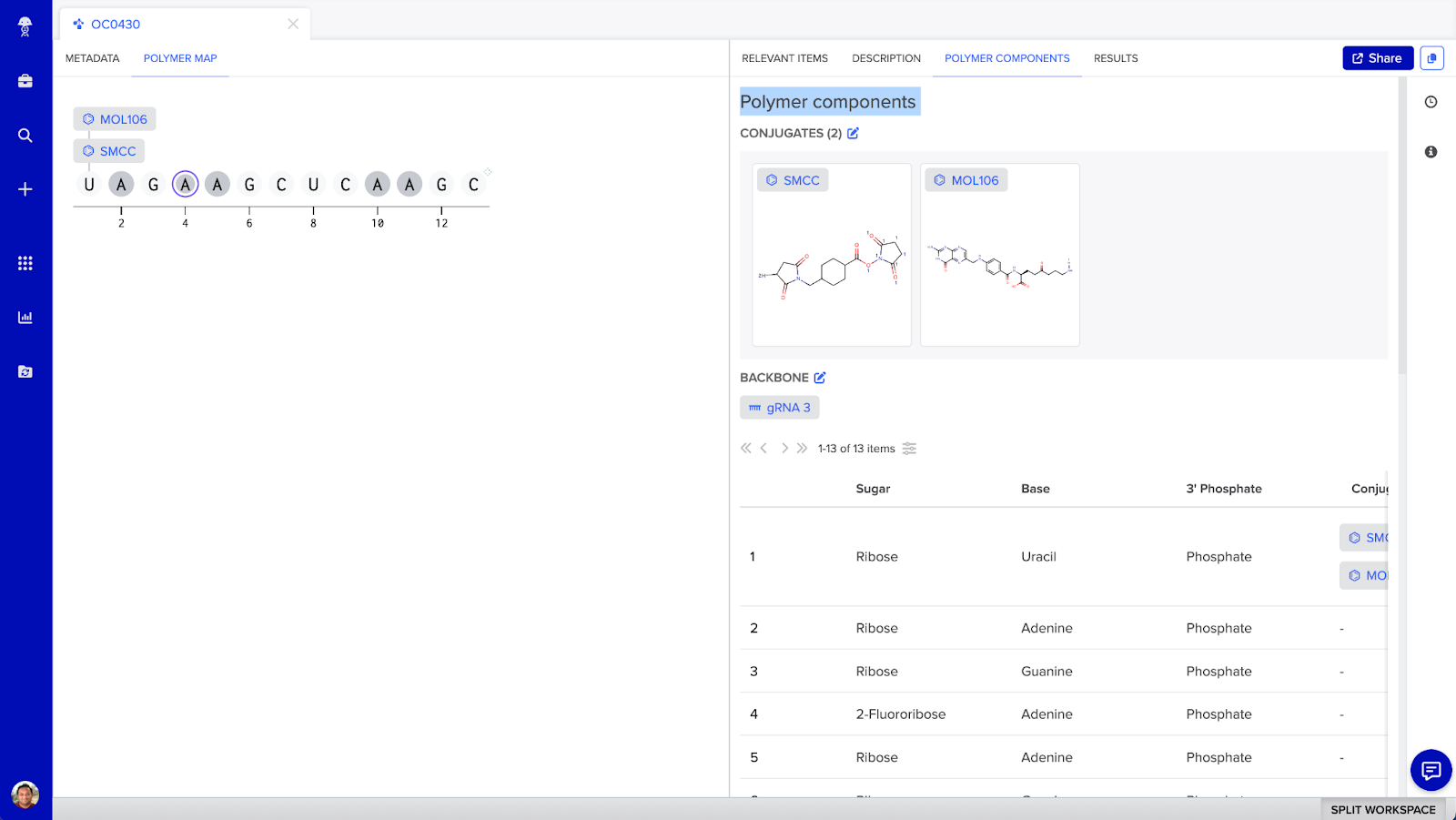
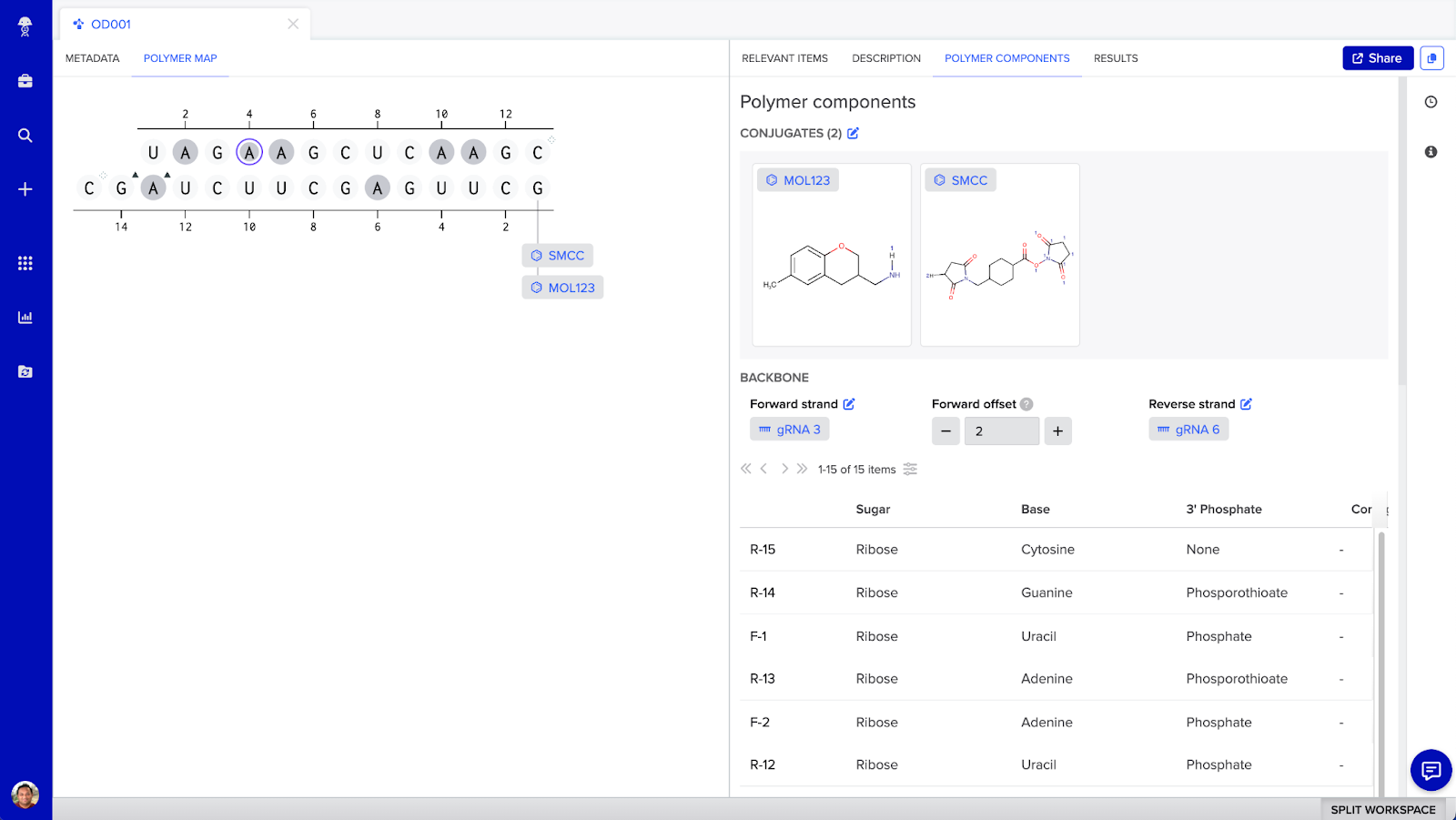
In Benchling you can visualize and design siRNA and ASO conjugates in a structurally accurate manner, bringing together multiple entity types. In addition to surfacing sequence level and structural information, scientists can define the discrete attachment points between multiple entities.
Here’s what’s included today
New oligo conjugate and oligo duplex entities to model and register ASO and siRNA conjugates
Visual map for oligonucleotide conjugates (such as ASOs) that are composed of an oligo entity and one or more molecules
Visual map for oligonucleotide duplexes (such as siRNA) that are composed of two oligo entities and one or more molecules
Ability to specify attachments between oligonucleotides and one or more molecules
Molecular properties calculation such as molecular weight and HELM notation
Auto linking of experiments and results with entity metadata
Oligonucleotide conjugate visual map
siRNA conjugate visual map
How modeling siRNA and oligonucleotide conjugates in Benchling helps you
Higher quality design and data: You can improve accuracy in visual representation of siRNA and oligonucleotide conjugates, leading to higher quality structural information and a reduction in errors during design.
Improved efficiency across team: You can ensure consistency of structural design across team members, leading to less duplication and lower user variability.
Faster research: The new entity types combined with the structured data model and unified platform of Benchling R&D Cloud help you aggregate experiments and results faster and make lead candidate selection or optimization decisions quickly.
References
GlobalData, accessed June 2024.
Delivery of oligonucleotide‐based therapeutics: challenges and opportunities, doi: 10.15252/emmm.202013243
Therapeutic siRNA: State-of-the-Art and Future Perspectives, doi: 10.1007/s40259-022-00549-3
Chemistry, structure and function of approved oligonucleotide therapeutics, doi: 10.1093/nar/gkad067
Therapeutic siRNA: state of the art, doi: 10.1038/s41392-020-0207-x
Therapeutic Oligonucleotides: An Outlook on Chemical Strategies to Improve Endosomal Trafficking, doi: 10.3390/cells12182253
Chemistry of Peptide-Oligonucleotide Conjugates: A Review, doi: 10.3390/molecules26175420
HELM: A Hierarchical Notation Language for Complex Biomolecule Structure Representation, doi: 10.1021/ci3001925
Learn more about Benchling for RNA therapeutics.
Powering breakthroughs for over 1,300 biotechnology companies, from startups to Fortune 500s