Benchling’s Codon Optimization Tool for Improved Protein Expression
Benchling offers a comprehensive suite of molecular biology tools that evolve with the latest life science research. Today, we are excited to add a brand new tool to that repertoire: Codon Optimization!
Researchers often try to maximize the expression of recombinant proteins in various host organisms. Protein expression depends on transfer RNAs (tRNAs), which translate a specific gene sequence to its corresponding amino acid sequence. Due to codon degeneracy, several tRNAs carry the same amino acid, resulting in 64 different combinations of codons for only 22 amino acids. Of the tRNAs that carry the same amino acid, some are more abundant than others. The tRNAs that are more abundant — and the degree by which they are — vary with each host organism. As a result, species-specific codon usage is an important factor to consider for improving protein expression. In many cases, optimizing the sequence of codons to systematically utilize abundant tRNAs during translation has significantly improved the yield of useful proteins.
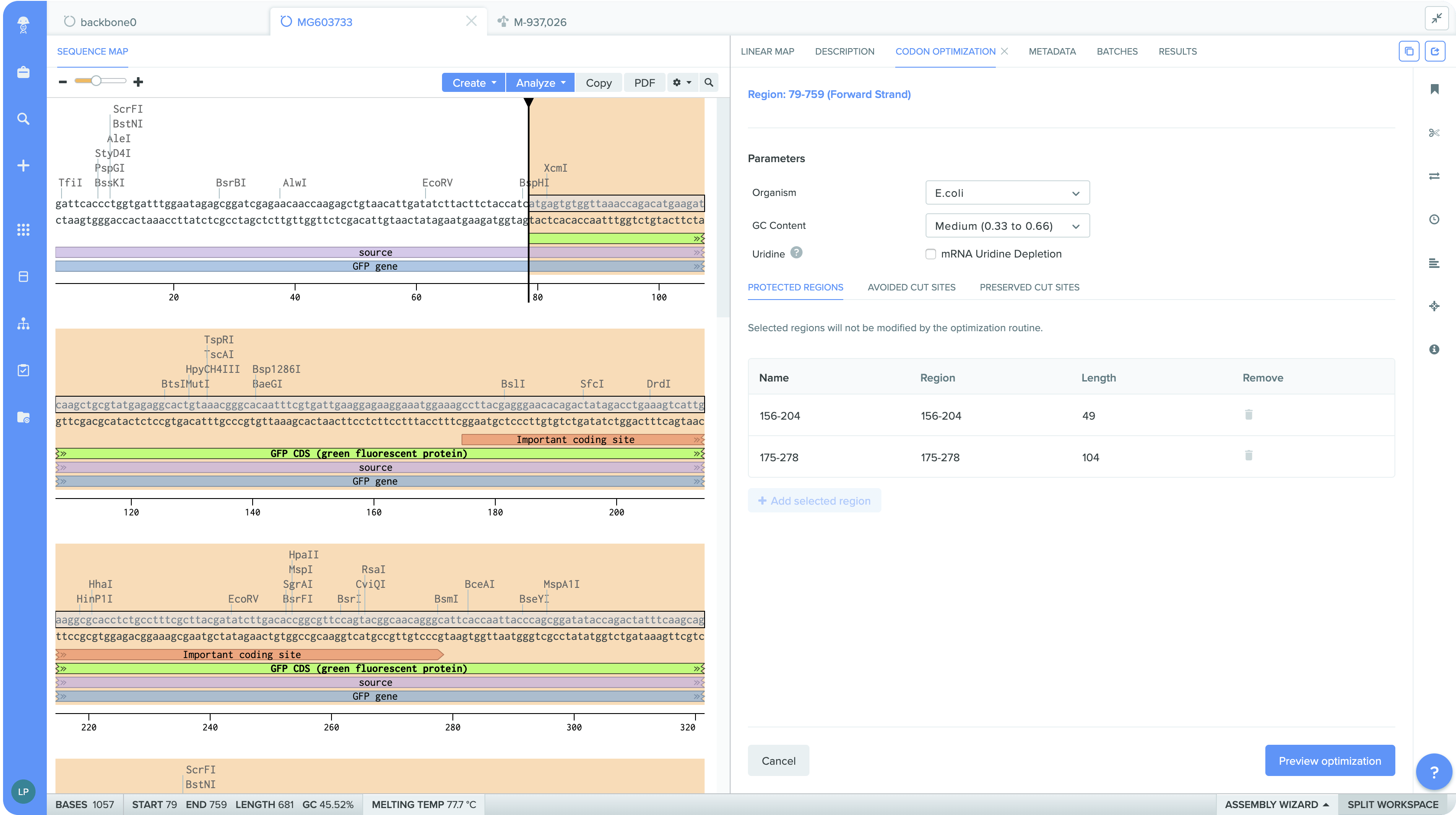
In Benchling, Codon Optimization streamlines the process of designing a DNA sequence to optimize protein expression in a target organism. Our new tool considers not only codon frequencies, but also GC content and other advanced parameters. In addition, this intuitive tool can perform optimized back translations of amino acid sequences.
Hear What Experts Are Saying: Watch the Webinar
To learn how to use our new Codon Optimization tool, watch our webinar, Codon Optimization with Benchling: Enhance Your Sequence Design and Protein Expression Workflows. This webinar showcases how this tool, integrated with our Molecular Biology application and data management platform, can help you increase protein expression and augment your research processes.
Enhance Your Protein Expression Workflows with Codon Optimization
Benchling’s Codon Optimization tool offers advanced features for customizing sequences and interpreting results. These features include the following:
Codon optimization based on an open source algorithm
Codon frequencies in a new sequence harmonize with the codon frequencies of the target organism
Support for back translation
Wide selection of common target organisms
Advanced input parameters, including GC content and mRNA uridine depletion
Option to conserve specific sequence regions, such as restriction sites and reserved sequences
Option to identify restriction sites that should not be generated by the optimization algorithm
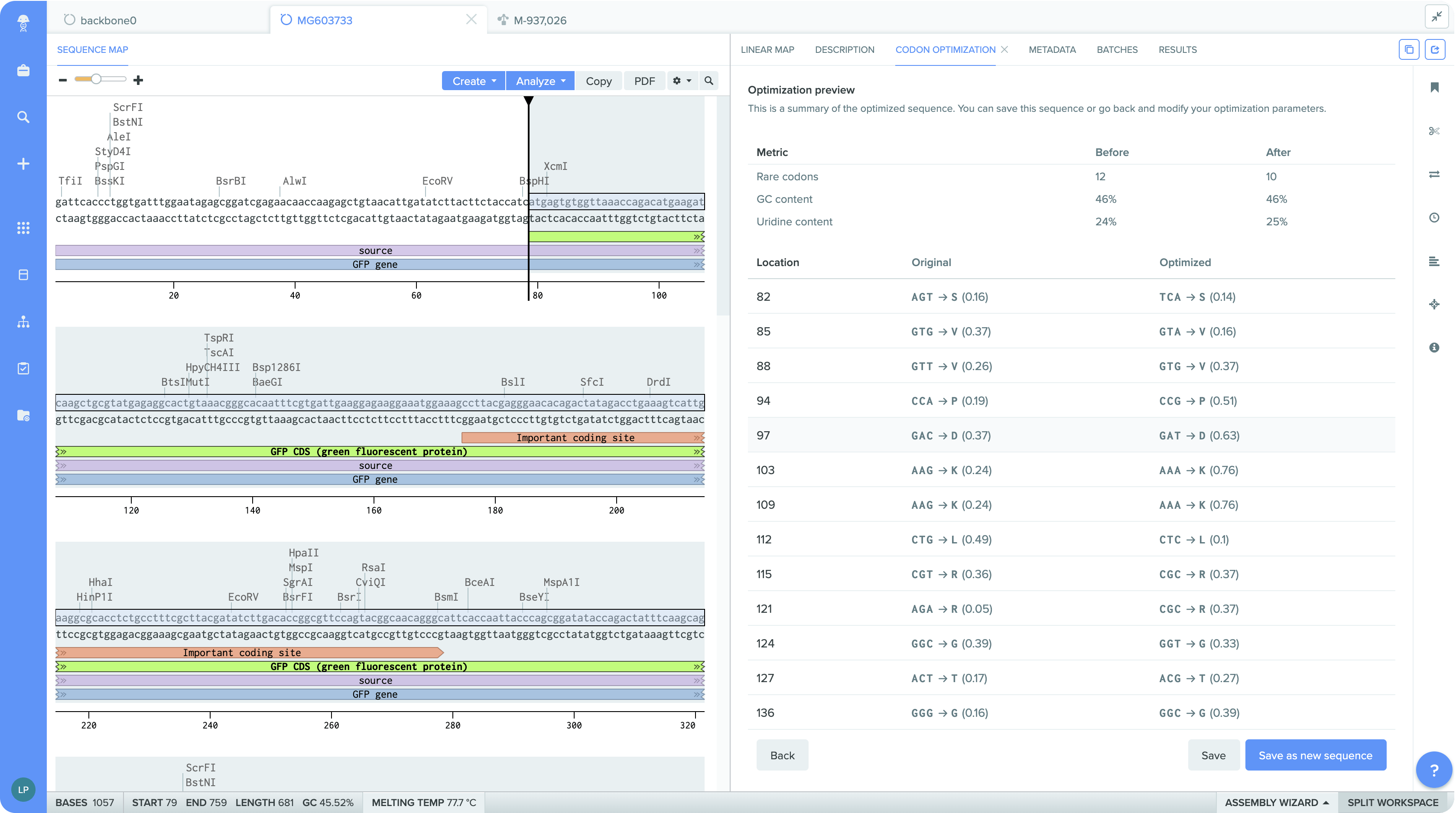
Easy-to-understand, transparent summary of changes to your sequence
Ability to tie expression data back to optimized sequences, with Structured Results in the Notebook
With all of these features, our Codon Optimization tool offers unparalleled benefits:
Trusted algorithm:Bring the relative frequencies of codons in your optimized sequences as close as possible to the frequencies found naturally in the target organism.
Advanced computational methodology: Get high-quality results by balancing codon prevalence with other important input parameters.
Powerful customization:Fine-tune experimental variables and expression levels by controlling GC content, uridine depletion, conserved sequences, and sequence avoidance.
Unified with Benchling Molecular Biology:Codon optimize your sequences and assemble them directly into plasmids, all within Benchling.
Ease of use:Get your team up and running with the new tool in a matter of minutes.
To learn more about our Codon Optimization tool, check out this product tutorial. To see the feature in action and get your questions answered, check out our webinar, Codon Optimization with Benchling.
Powering breakthroughs for over 1,200 biotechnology companies, from startups to Fortune 500s



