Generating FAIR — Findable, Accessible, Interoperable, Reusable — Resources with Marchantia, a Prototype for Plant Synthetic Biology
Among scientists, the COVID-19 crisis has clarified the importance of the ability to easily share scientific data among collaborators, or even the community overall. In order to rapidly utilize others' datasets, scientists need to ensure that their data is accessible, standardized, and contextualized. In this article, Dr. Susana Sauret-Gūeto, a biologist at the University of Cambridge, explains how she built a culture in her lab that prioritized capturing and sharing well-characterized data. Though her work is in plant biology, not infectious disease, the lessons she conveys are universally powerful — and are especially applicable to the current circumstances.
“Classic!” That's a veteran lab member's response when a new PhD student recognizes that a plasmid doesn't have the expected sequence.I have personally heard this, and I have seen it happening around me many more times. DNA constructs from other labs, or even from former members of your own lab, can often be poorly documented. As a result, scientists waste time and resources by having to recheck or redo previous work. And that’s just the tip of the iceberg when it comes to data management challenges in a highly dynamic and resource-limited academic environment. In this environment, working practices and structural constraints have tended to prioritize innovation before documentation. Fortunately, this has been changing for some time, as data-driven biology and society have demanded well-characterized research and open science in order to make research more reproducible and more transparent.

This article was contributed by:
Dr. Susana Sauret-Gūeto
— Dr. Susana Sauret-Gūeto is a Research Manager at OpenPlant in the Department of Plant Sciences and a Data Champion at the University of Cambridge.
FAIR Data in the Field of Synthetic Biology
When I joined OpenPlant to work on Marchantia as a prototype for plant synthetic biology, we were planning on generating and testing hundreds of DNA constructs as part of a collaborative project. I saw an opportunity to influence good data management practices in the lab and to help facilitate the production of FAIR data. FAIR stands for Findable, Accessible, Interoperable, and Reusable. Generating FAIR data requires a research culture that values collaborative work and recognizes the need for data stewards, data curators, and data management tools to assist in the research workflows. Conveniently, the field of synthetic biology brings an engineering perspective to biology and not only broadens the space of applications, but also triggers operational changes1 as it promotes standardization and collaboration. To deliver on the promises of synthetic biology, it is essential to learn from and reuse well-characterized data to iterate through the design-build-test-learn cycle. Even so, the generation of FAIR data should go beyond producing accessible and well-documented data registries. Generating FAIR data should involve the adoption of lab practices to easily document essential metadata as it is generated, in a way that benefits researchers in their day-to-day work.2
OpenPlant: A Plant Synthetic Biology Initiative
OpenPlant is a research initiative part of the UK Synthetic Biology for Growth Programme that brings together researchers from the University of Cambridge and two institutions in Norwich, UK: the John Innes Centre and the Earlham Institute. OpenPlant's main goals are to facilitate interdisciplinary exchange, development of new tools and methods for plant synthetic biology, open sharing of standardized resources, and responsible innovation for improvement of sustainable agriculture and conservation. Plant synthetic biology approaches, like ours, are especially important in the face of global threats from new pathogens, climate change, soil degradation, restricted land use, salinity, and drought.
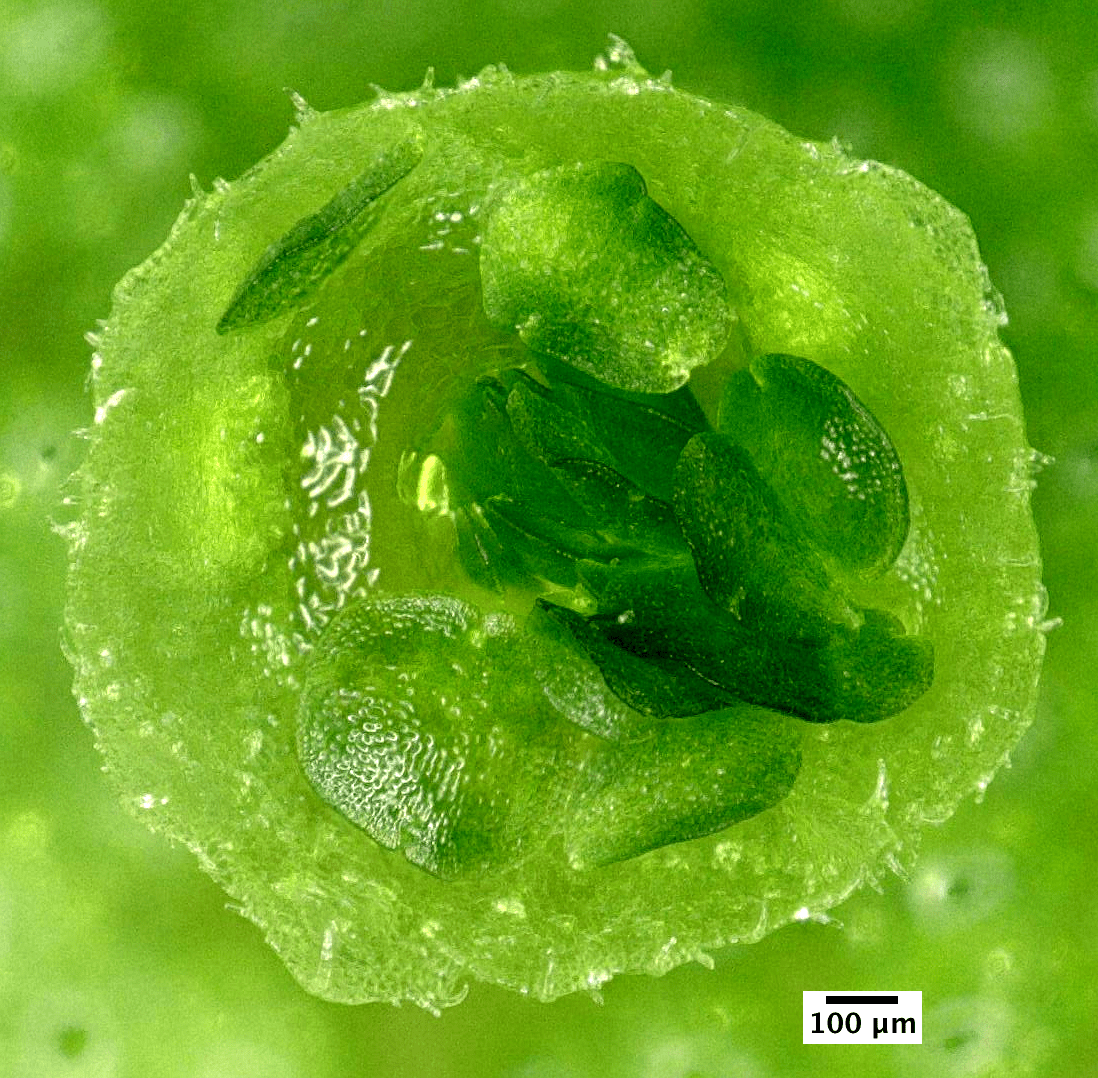
A Gemmae are clonal propagules that, once detached from the parent plant and dispersed, grow into a new Marchantia plant. Gemmae development happens in the open, allowing easy observation of morphogenesis processes with microscopy. The width of mature gemmae inside the gemmae cup is about half a millimeter.
Plants and algae are attractive platforms for synthetic biology as they are photoautotrophs, can potentially be grown at large scale, and have extensive secondary metabolism. Chloroplasts, the photosynthetic organelles in plants and algae, have a simple and prokaryote-like genome, with the ability to support high levels of transgene expression, making them amenable to synthetic biology approaches.3,4 Plants grow through iteration of modules and are plastic, adapting their growth and development to the environment. This modularity and plasticity makes plants a unique platform to address challenging questions on the self-organization of all multicellular organisms. In plants, synthetic biology could allow the production in specific cell types of familiar or new-to-nature compounds for food, medicine, or industry, like human proteins being produced in the moss Physcomitrella patens.5 It could also allow the manipulation of familiar morphological structures,6 like the “smart” domestication of fruits.7
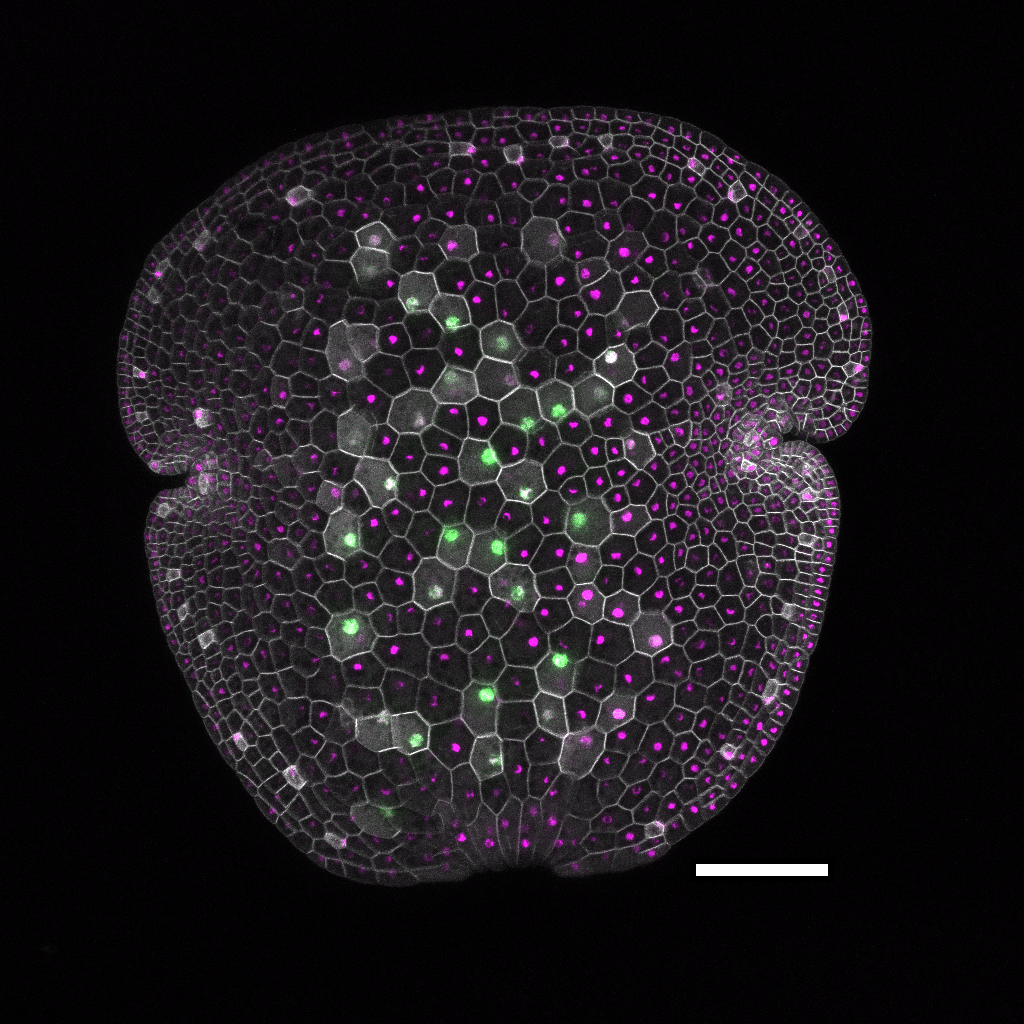
Construction of precise markers for visualizing dynamic changes in expression patterns — to advance the understanding of morphogenesis processes in Gene assemblies for multispectral imaging of live gemmae are built with Loop DNA assembly and the OpenPlant toolkit. This image shows a line expressing a set of marker genes: p5-MpRSL3:mVenus-N7 (nuclear-localized rhizoid precursor and rhizoid cell marker, green), p5-MpUBE2:mTurquoise2-N7 (nuclear-localized ubiquitous expression, magenta) and p5-MpUBE2:mScarletI-LTI6b (plasma membrane-localized ubiquitous expression, grey). Maximum intensity z-axis projection image of a gemma just removed from the gemmae cup (0 day). Scale bar: 100 μm. (More information in Sauret-Gueto, et al.11)
Simple and rapid prototype plant systems are needed to complement the work on model vascular plants and crops, like Arabidopsis, tomato, or wheat, which have relatively long generation times, functional redundancy in diploid or more complex genomes, and difficult access to early stages of organ formation. This is why, in the OpenPlant Synthetic Biology and Reprogramming of Plant Systems group (the Haseloff Lab) at the Department of Plant Sciences, University of Cambridge, we are working with the liverwort Marchantia polymorpha. Marchantia is a morphologically simple multicellular plant with a fast life cycle and a dominant haploid phase that can easily be transformed.8,9,10 The development of its plantlets happens in the open, allowing easy observation of morphogenesis. Marchantia also has a small genome size with low genetic redundancy.10 It shares most of the characterized gene signalling components with more recently diverged land plants, like Arabidopsis, but has fewer genes in each gene family.
We are interested in using a simple plant testbed to rewire signalling networks controlling tissue architecture and growth, and to engineer the production of compounds in specific cell types and organelles, particularly the chloroplast (Figure 1). We expect the use of Marchantia will help uncouple the links between genetic networks, cell autonomous and non-autonomous processes, and tissue-wide physical processes that drive morphogenesis. This work will be useful for the synthetic biology community working on self-organization of multicellularity, as well as the plant research community studying development and the evolution of signalling pathways in plants.
Developing the OpenPlant Toolkit
Our first aim has been to expand the tools and resources to work with Marchantia.11 We have adopted techniques for simple and efficient propagation and maintenance of Marchantia lines throughout its life-cycle, with no requirement for specialized glasshouse facilities (Figure 1). Former and current researchers in the lab have developed modular and standardized DNA assembly methods and created registries of DNA parts and devices (Figure 1). We have also adopted multiwell plates for culture and for the screening of multiple lines under microscopic observation. To ensure that our work is open and accessible, we have made the lab's Marchantia protocols available through an OpenPlant project on protocols.io.
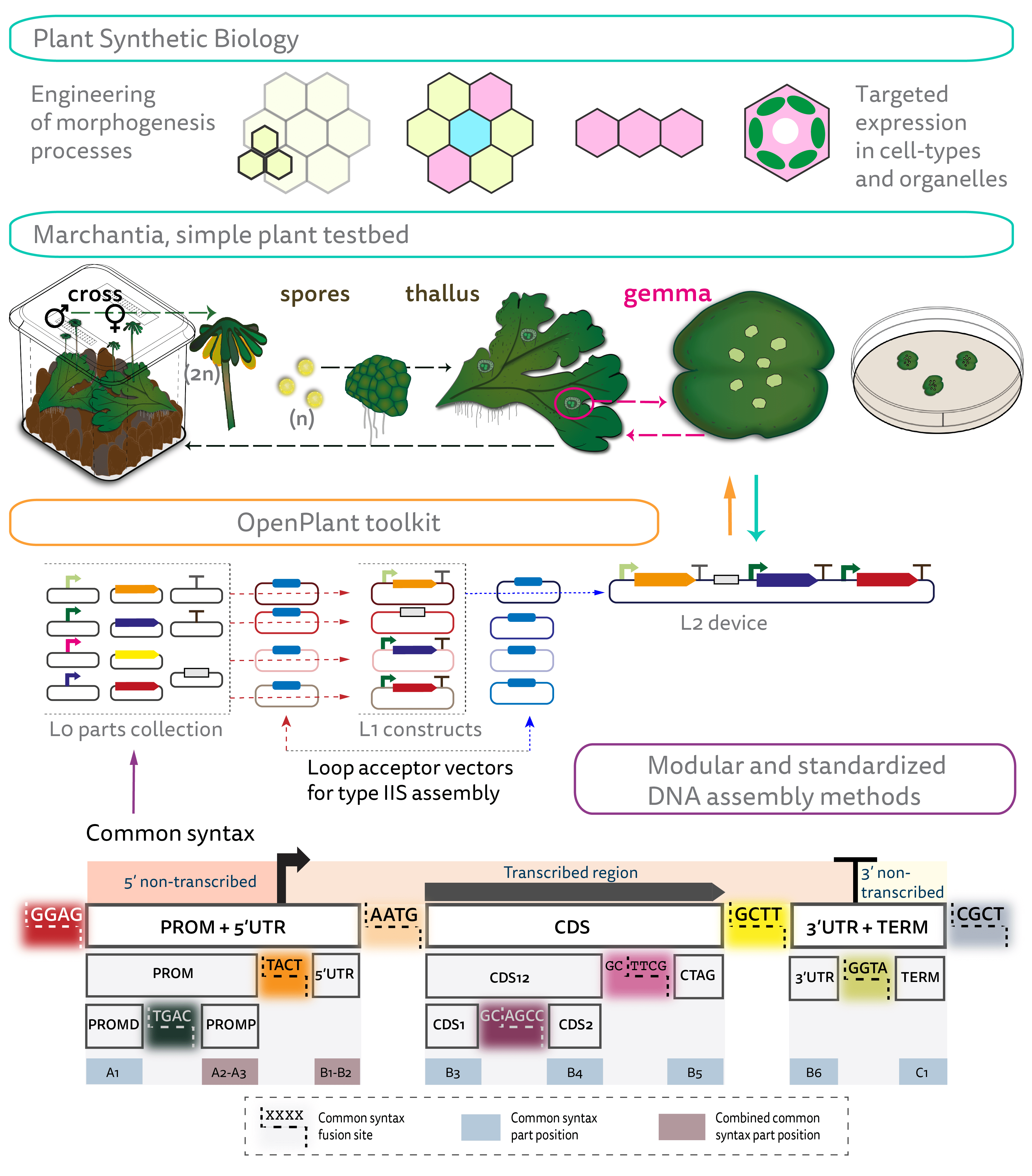
Figure 1. Engineering approaches in plant synthetic biology with a simple plant testbed, Plant synthetic biology offers the prospect to reprogramme plant form and function. Marchantia is a morphologically simple plant with a dominant haploid phase and a fast life-cycle. It propagates sexually through spores and asexually through gemmae and can be easily maintained in sterile plates and boxes without requirement for glasshouse facilities. Type IIS DNA assembly methods, like Loop, allow for modular, hierarchical and standardized generation of DNA devices, assembling Level (L0) parts into transcriptional units (Level 1, L1) and L1 constructs into Level 2 (L2) devices. The OpenPlant toolkit contains the Loop vectors and a collection of Level 0 parts to use in Marchantia for nuclear and chloroplast transformation and CRISPR genome editing. The common syntax breaks down a transcriptional unit into modular parts (L0 parts) with specific fusion sites. Examples of L0 parts are PROM+5UTR, CDS or 3UTR+TERM. This allows combinatorial assembly of collections of level 0 parts. (Adapted from Sauret-Gueto, et al.11)
To test and analyze large numbers of plants transformed with different variants of DNA devices, we first needed an organized, modular system that would enable us to easily assemble multiple devices from a pool of basic DNA parts. In OpenPlant, we follow the common syntax, agreed upon by the community, to break down a transcription unit into standardized basic DNA parts called Level 0 (L0) parts and to define specific fusion sites between each type of L0 part (Figure 1). Complimentary fusion sites between L0 parts determine the order of assembly of the parts. We use type IIS cloning for DNA assembly because the type IIS recognition sites are lost during the assembly, giving a direction to the reaction and allowing for a digestion-ligation one-pot reaction that is highly efficient.12,13 The efficiency of the type IIS assembly allows us to scale up and even automate the production of DNA devices.11,14
In particular, we are using the Loop type IIS assembly method to assemble L0 parts into transcription units (Level 1, L1) and L1 constructs into devices (Level 2, L2)11,15 (Figure 1). To facilitate work with Marchantia, we have developed the OpenPlant toolkit, which includes Loop nuclear transformation vectors, Loop vectors for chloroplast transformation and Loop vectors for CRISPR genome editing. The toolkit also contains a collection of standardized L0 parts that have been tested in multiple devices for expression in Marchantia, including antibiotic resistance genes, signal peptides, fluorescent proteins for multispectral imaging and promoters for gene expression.11
A Pipeline that Promotes Collaboration, Transparency, and the Generation of Standardized, Reusable Data
In the lab, the generation of multiple DNA devices and Marchantia lines has been a team effort. This effort is facilitated by tools that enable good data management practices. We have adopted Benchling because it provides an electronic notebook, molecular biology suite, and collaborative data management platform. Benchling has an Assembly Wizard, which we use to model assembly of the final plasmid, while checking the cloning strategy and tracing the plasmid lineage. It also supports the development of lab-tailored registries and custom fields associated with particular types of plasmids. We have established a pipeline in the lab that allows all of the members involved in a cloning project to work together, organizing the inventory of DNA constructs by groups of L0, L1, or L2 constructs. Once constructs are finalized, they are moved to a lab Registry folder, given a unique ID, and checked for annotations and metadata (Figure 2).
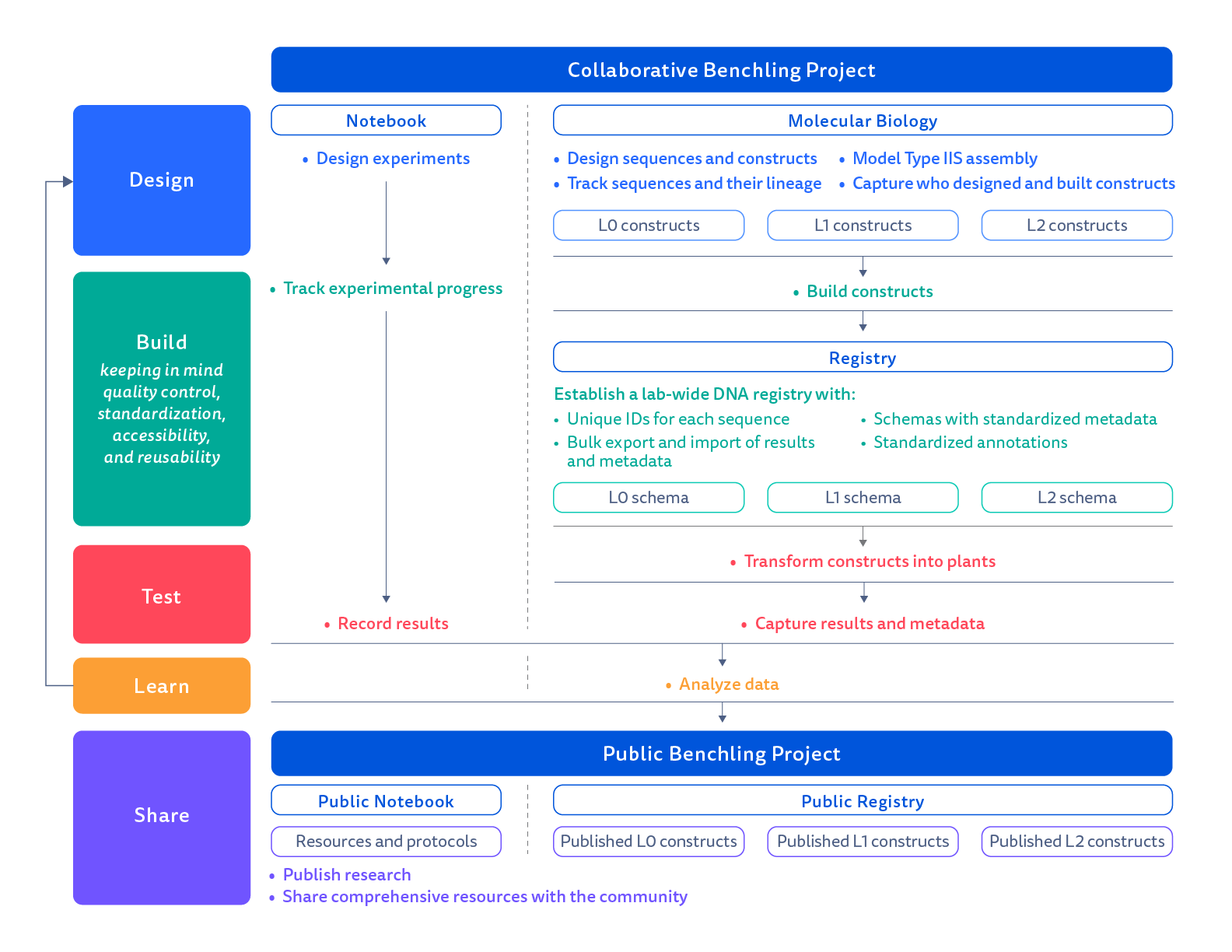
Figure 2. Pipeline for data management using Benchling. Benchling provides a collaborative data management platform to support the design-build-test-learn cycle. The workflow of Marchantia experiments in the lab starts with a collaborative Benchling project that includes an electronic notebook and an inventory of type IIS constructs. Once built, we move constructs into a lab Registry. And once we test the constructs in Marchantia, analyze the results, and are ready to share the research project with the community, we move information on our constructs into a public Benchling project.
The pipeline facilitates teamwork and ensures that data is accessible and reusable: it maintains reliable information on constructs generated by researchers who are no longer in the lab, allows new researchers to easily find information on lab constructs, facilitates standardization of annotations and metadata, and provides an electronic notebook to track experimental details and progress. All of this supports the scaling up of experiments and the generation of reproducible datasets. Moreover, once a research project is ready for publication and sharing with the research community, Benchling's academic product gives users the opportunity to copy all relevant DNA constructs to a public folder, supporting accessibility by the community and reproducibility in science.
The OpenPlant toolkit is being deposited in Addgene for global distribution under an OpenMTA licence. Use of the OpenMTA places the materials into the public domain, allowing their redistribution and commercial use. We have also shared the information on the vectors and parts through a public Benchling folder. Already, the kit has been well received by the Marchantia community; of special interest has been the identification of a new promoter for ubiquitous expression and a new promoter for specific expression in the rhizoids, simple root cells in Marchantia.11
We expect the OpenPlant toolkit, together with simplified methods for propagation, transformation and observation of Marchantia lines, to facilitate work with Marchantia for basic research. Not only that, it will also lower the logistical threshold for plant experiments and promote the adoption of this plant system by the broad synthetic biology community. We hope the modular and standardized nature of the collection will galvanize the production and interchange of further collections of DNA parts. For instance, we are using the OpenPlant toolkit to identify tissue-specific promoters by building a collection of devices with the proximal promoters of the ~400 Marchantia transcription factors,10 screening the transformed lines through fluorescence microscopy.
Building a Culture of Strong Data Management
Going back to my journey to influence good data management practices in the lab, my work at OpenPlant developing tools for Marchantia, promoting standardization practices, and the production of well-characterized data that is findable and reusable, has been facilitated by data management tools like Benchling and Rstudio (which I did not talk about in this post). From my experience, useful tools are driven by the right people. Benchling has been successful in the lab because the group of Marchantia researchers are people who see the benefit of teamwork and the benefits of Benchling's tools for their individual and collaborative projects. Working with a supportive group has greatly facilitated my work coordinating research projects and curating lab data.
During my time in OpenPlant, I established the ROC (Researchers with OpenPlant in Cambridge) group, putting in contact many researchers who see the importance of producing FAIR data. These individuals need to be supported by the organization's culture. In this sense, the data champion initiative at the University of Cambridge, is a first step in raising awareness of good data management practices. And at some universities, like Delf University of Technology in the Netherlands, they have already recognized the need for data stewards to support research data management across campus. Academic research is starting to adapt to support standardization, generation of FAIR data, sharing of information, reproducibility and innovation in the data-driven biology era. Alleluya!
Acknowledgements
I would like to thank Professor Jim Haseoff, Director of OpenPlant and Principal Investigator of the Synthetic Biology and Reprogramming of Plant Systems group at the Department of Plant Sciences, University of Cambridge. I would also like to thank all past and present lab members part of the Marchantia team — especially Eftychios Frangedakis, Marta Tomaselli, Marius Rebmann, Alan Marron, Jenna Rever, and Linda Silvestri — and all past members of the ROC group — Steven Burgess, Payam Mehrshahi, Katrin Geisler, Francisco Navarro, Chiara Airoldi and Jan Lyczakowski to name a few. Thanks to Jenny Molloy and Marta Teperek for discussions on FAIR data. And finally, thanks to Lily Helfrich and Steven Burgess for feedback on the post.
References
Sauret-Gueto, S. et al.Systematic tools for reprogramming plant gene expression in a simple model, Marchantiapolymorpha. doi:10.1101/2020.02.29.971002.(BioRxiv)
Join Susana in generating standardized, accessible data on Benchling: